Abstract
Leaf elongation rate (LER) is an important factor controlling plant growth and productivity. The objective of this study was to determine whether genetic variation in LER for a fast-growing (‘K-31’), and a dwarf cultivar (‘Bonsai’) of tall fescue (Festuca arundinacea) and gibberellic acid (GA) regulation of LER were associated with differential expression of cell-expansion genes. Plants were treated with GA3, trinexapac-ethyl (TE) (GA inhibitor), or water (untreated control) in a hydroponic system. LER of ‘K-31’ was 63% greater than that of ‘Bonsai’, which corresponded with 32% higher endogenous GA4 content in leaf and greater cell elongation and production rates under the untreated control condition. Exogenous application of GA3 significantly enhanced LER while TE treatment inhibited leaf elongation due to GA3-stimulation or TE-inhibition of cell elongation and production rate in leaves for both cultivars. Real-time quantitative polymerase chain reaction analysis revealed that three α-expansins, one β-expansin, and three xyloglucan endotransglycosylase (XET) genes were associated with GA-stimulation of leaf elongation, of which, the differential expression of EXPA4 and EXPA7 was related to the genotypic variation in LER of two cultivars. Those differentially-expressed expansin and XET genes could play major roles in genetic variation and GA-regulated leaf elongation in tall fescue.
Similar content being viewed by others
Introduction
Leaf growth is the major determining factor contributing to shoot biomass and yield production1,2. Leaf elongation rate (LER) is genetically controlled and developmentally regulated and varies with leaf age, leaf position, and between fast-growing cultivars and slow-growing cultivars3,4, but it is also very sensitive to external factors5,6,7. Depending on the objectives of plant production, either fast-growing or slow-growing leaves may be desirable, selectable traits in plant improvement. For example, for perennial grass species, fast-growing species are desirable for the productivity of grasses in forage or natural grasslands while slow-growing traits are important for turf grasses requiring mowing8,9. Therefore, understanding the mechanisms controlling leaf elongation is critically important for genetic modification of plants for fast- or slow-growing habits through transformation or molecular breeding.
Leaf elongation is controlled by cell elongation and cell division rates10,11. Both of those processes are located in the base of the elongating leaf which is called the leaf elongation zone and enclosed by the sheaths of older leaves in grasses12. The relative importance of each cell process accounting for the variations in leaf elongation rate is also variable, depending on plant species and environmental factors. The LER may be determined by both of cell elongation and production rates in some grass species, such as tall fescue (Festuca arundinacea), while the variations in LER can also be due to mostly differences in cell production rate rather than cell elongation in some other grass species, such as Poa compressa, P. annua, and P. trivialis13. Volenec et al.14 found a high-LER cultivar (26 mm d−1) of tall fescue had 25% longer epidermal cells and 24% higher cell production rate than a low-LER cultivar (18 mm d−1), suggesting genotypic variation in LER related to both cell elongation and production rate14. The cellular and molecular factors accounting for the genetic variation in leaf elongation are still not well understood.
Cell elongation is controlled by cell extensibility, which is regulated by cell-wall loosening proteins and enzymes including expansin and xyloglucan endotransglycosylase (XET)15,16. Expansin is the primary factor in the cell wall that mediates pH-dependent wall loosening, which can disrupt the non-covalent binding between the cell wall polysaccharides, thereby allowing turgor-driven wall extension17. Expansins are encoded by a large gene family including two major types of α-expansins (EXPA) and β-expansins (EXPB)18,19, which play critical roles in regulating cell expansion in the leaf elongation zone20. Previous studies have identified many expansin genes in maize (Zea mays)21, rice (Oryza sativa)22, wheat (Triticum aestivum)23, meadow fescue (F. pratensis)24 and other species25,26,27, the expression levels of which were positively related to leaf and stem elongation. Goh et al.28 reduced the expression of several expansin genes in arabidopsis (Arabidopsis thaliana) using an inducible microRNA construct and found that the decreased expansin gene expression led to a repression of leaf growth28. Over-expression of an expansin gene cloned from rice (OsEXPA4) increased the coleoptile and mesocotyl length by up to 31 and 97%, respectively, while in the anti-sense seedlings, the length of them decreased by up to 28 and 43%, respectively29.
XET is another important protein located in cell wall that has been associated with cell elongation in various plant species30. Many previous studies demonstrated that XET activity and gene expression were positively correlated to the elongation rate of leaf blades in grasses, such as barley (Hordeum vulgare)31, meadow fescue24, and maize32. There are various members of expansins and XET genes, but the specific genes related to genetic variation in leaf elongation are not well documented.
In addition, hormones, such as gibberellins (GAs), are known to affect leaf elongation in many species, such as arabidopsis33, maize34 and rice34,35. Most semi-dwarf cultivars with lower leaf growth rate were found to have either less sensitivity to GA or reduced levels of endogenous GA36,37. Bultynck and Lambers38 examined the effects of GA3 and paclobutrazol, an inhibitor of GA biosynthesis, on two Aegilops species with contrasting leaf elongation rates and found that addition of GA3 increased leaf elongation rate of both Aegilops species via stimulating both cell elongation and division while paclobutrazol inhibited leaf elongation rate via repressing cell elongation and division38. Similar results were also reported in wheat39 and barley40. However, whether genetic variation and the effects of GA on the elongation of leaves are associated with changes in expansin and XET expression is not clear. Understanding cellular and molecular mechanisms underlying genetic variations and hormonal regulation of leaf elongation will provide further insights into strategies to develop plants with desirable traits of fast-growing or slow-growing phenotypes.
Tall fescue has wide genetic variation in leaf elongation rate, with cultivars of fast-growing or slow-growing (or dwarf-type) phenotypes widely used as forage and turf grasses, respectively41,42. The various growth habits make tall fescue a good model species for studying mechanisms controlling leaf elongation in perennial grasses. In this study, it is hypothesized that the genetic variation in leaf elongation between fast-growing and dwarf-type tall fescue cultivars could be regulated by differential responses to GA, endogenous production of GA, and/or differential expression of cell-wall loosening genes controlling cell elongation. Therefore, the objectives of this study were to determine GA-regulation of leaf elongation and differential expression of several expansin and XET genes associated with the genetic variations in leaf elongation rate by comparing a fast-growing cultivar ‘K-31’ and a dwarf-type cultivar ‘Bonsai’.
Results
Differential leaf elongation rate between cultivars
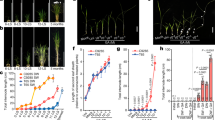
Leaves of ‘K-31’ and ‘Bonsai’ exhibited differential elongation rate, and the differences became more pronounced with leaf age. The first leaf elongation rate of ‘K-31’ (10.52 mm d−1) was 19% higher than ‘Bonsai’ (8.82 mm d−1) (Fig. 1A–C); the second leaf elongation rate of ‘K-31’ (16.34 mm d−1) was 48% greater than ‘Bonsai’ (11.06 mm d−1) (Fig. 2A–C); and the third leaf was 57% greater in ‘K-31’ (20.09 mm d−1) than ‘Bonsai’ (12.77 mm d−1) (Fig. 3A–C).
(A) The first leaf length of both cultivars in the elongating phase during 12-d emergence. The vertical bar is the standard error of mean leaf length (n = 40 replicates) at each given day of leaf emergence. (B) Changes of the first leaf length during the linear growth phase within the first 4 d of leaf emergence for ‘Bonsai’. (C) Changes of the first leaf length during the linear growth phase within the first 5 d of leaf emergence for ‘K-31’. The slope of the linear regression line represents leaf elongation rate (mm d−1) in (B) and (C). The function y = mx + b represents the linear relationship of leaf length (y) to days of leaf elongation (x) and the LER (m) was calculated by the equation m = [n∑(xy) − ∑x∑y]/[n∑(x2) − (∑x)2]. The R2 is the square of the correlation coefficient.
(A) The second leaf length of both cultivars in the elongating phase during 12-d emergence. The vertical bar is the standard error of mean leaf length (n = 40 replicates) at each given day of leaf emergence. (B) Changes of the second leaf length during the linear growth phase within the first 5 d of leaf emergence for ‘Bonsai’. (C) Changes of the second leaf length during the linear growth phase within the first 6 d of leaf emergence for ‘K-31’. The slope of the linear regression line represents leaf elongation rate (mm d−1) in (B) and (C). The function y = mx + b represents the linear relationship of leaf length (y) to days of leaf elongation (x) and the LER (m) was calculated by the equation m = [n∑(xy) − ∑x∑y]/[n∑(x2) − (∑x)2]. The R2 is the square of the correlation coefficient.
(A) The third leaf length of both cultivars in the elongating phase during 12-d emergence. The vertical bar is the standard error of mean leaf length (n = 40 replicates) at each given day of leaf emergence. (B) Changes of the third leaf length during the linear growth phase within the first 5 d of leaf emergence for ‘Bonsai’. (C) Changes of the third leaf length during the linear growth phase within the first 6 d of leaf emergence for ‘K-31’. The slope of the linear regression line represents leaf elongation rate (mm d−1) in (B) and (C). The function y = mx + b represents the linear relationship of leaf length (y) to days of leaf elongation (x) and the LER (m) was calculated by the equation m = [n∑(xy) − ∑x∑y]/[n∑(x2) − (∑x)2]. The R2 is the square of the correlation coefficient.
The REGR along the third leaf was compared between the two cultivars (Fig. 4). The maximum REGR of ‘K-31’ was 14% higher than ‘Bonsai’. The length of elongation zone was also longer in ‘K-31’ compared with ‘Bonsai’, as ‘Bonsai’ leaf reached to the maximum elongation rate within 6 mm from the leaf base while ‘K-31’ leaves did not increase to the peak rate until 10 mm from the leaf base and maintained significantly greater rate than ‘Bonsai’ beyond 10 mm from the leaf base.
Cultivar variations and exogenous GA application in endogenous GA content
To investigate whether differences in LER could be related to GA levels, endogenous GA1 and GA4 contents of leaves were compared between the two cultivars with or without exogenous GA treatment. ‘K-31’ leaves had significantly higher endogenous GA4 level than ‘Bonsai’ leaves but there were no significantly differences in GA1 contents between those two genotypes (Fig. 5). The endogenous GA4 contents of leaves increased 3.77 fold and 1.64 fold by exogenous application of GA in ‘K-31’ and ‘Bonsai’, respectively. The endogenous GA1 content of leaves kept the same level in ‘Bonsai’ after GA application and increased by 54% in ‘K-31’.
Endogenous GA4 (A) and GA1 (B) content of the third leaves of ‘K-31’ and ‘Bonsai’ with and without GA (50 μmol L−1 GA3) application. The vertical bar is the standard error of mean (n = 4 replicates of GA treatment, and each replicate contains at least 10 seedlings). Columns marked with different letters indicate significant differences between treatments and between cultivars based on LSD test (P = 0.05).
Effects of exogenous GA and TE application on leaf elongation, cell elongation, and cell division
Exogenous application of GA significantly enhanced LER in both cultivars, with 61% and 66% greater leaf elongation rate in GA-treated leaves than untreated control leaves for ‘K-31’ and ‘Bonsai’, respectively (Fig. 6). In contrast, TE application inhibited leaf elongation in both cultivars, but to a greater extent for ‘Bonsai’ than ‘K-31’, with 31% reduction in leaf elongation rate of TE-treated ‘K-31’ and 60% reduction for ‘Bonsai’ (Fig. 6).
Leaf length (A) and leaf elongation rate (B) of the third leaf for ‘K-31’ and ‘Bonsai’ as affected by GA or TE treatment. The vertical bar is the standard error of mean leaf length (n = 4 replicates of GA or TE treatment, and each replicate contains at least 10 seedlings). Columns marked with different letters indicate significant differences between treatments and between cultivars based on LSD test (P = 0.05).
In order to determine whether enhanced leaf elongation was due to increases in cell length and/or increases in cell production rate, the length of two types of epidermal cells and cell production rate of each cell type were compared between two cultivars with or without GA and TE treatments. The epidermal long cells in ‘K-31’ were longer than that in ‘Bonsai’ either with or without GA or TE application (Fig. 7A). However, the length of interstomatal cells did not differ between the two cultivars (Fig. 7B). The application of GA resulted in significant increases in epidermal long cells and interstomatal cells for both cultivars while TE inhibited cell length of both cell types in both cultivars. The cell production rate of both types of epidermal cells was significantly greater in ‘K-31’ than that in ‘Bonsai’ regardless of GA or TE treatments (Fig. 8A,B). Application of GA increased the production rate of both types of epidermal cells for both cultivars whereas TE treatment resulted in significant reduction in cell production rate for both epidermal types in “Bonsai’ leaves and only interstomatal cell production rate in ‘K-31’ leaves.
(A) Long cell; (B) Interstomatal cell. The vertical bar is the standard error of mean (n = 4 replicate of GA or TE treatment, and each replicate contains at least 10 seedlings). Columns marked with different letters indicate significant differences between treatments and between cultivars based on LSD test (P = 0.05).
(A) Long cell; (B) Interstomatal cell. The vertical bar is the standard error of mean (n = 4 replicates of GA or TE treatment, and each replicate contains at least 10 seedlings). Columns marked with different letters indicate significant differences between treatments and between cultivars based on LSD test (P = 0.05).
Differential expression of expansin and XET between cultivars and responses to GA and TE treatment
In order to determine whether the genetic variations and GA effects on leaf elongation are due to differences in the expression level of genes regulating cell elongation, transcript levels of several expansins and XET were analyzed. Five expansin ESTs (EXPA4, EXPA5, EXPA7, EXPB4, EXPB7) and three XET ESTs (XET1, XET2, XET3) were identified in tall fescue through EST search in NCBI database. Among the 5 expansin genes, 4 expansins including three α-expansins (EXPA4, EXPA5 and EXPA7) and one β-expansin (EXPB4) were up-regulated by GA treatment in both cultivars, and only EXPB7 was down-regulated by TE treatment in ‘K-31’ (Figs 9 and 10). Compared between the two cultivars, the expression level of EXPA7 was significantly higher in ‘K-31’ than ‘Bonsai’ either with or without GA or TE treatment while EXPA4 showed consistence differences between the two cultivars exposed to control and GA treatment. EXPB7 and XET1 neither show responses to GA treatment nor exhibit cultivar variations (Figs 10 and 11). The expression level of XET2 and XET3 genes did not show consistent differences between the two cultivars exposed to control, GA or TE treatment, but they exhibited differential responses to GA and TE treatment in both cultivars (Fig. 11). XET1 and XET2 expression increased with GA treatment for ‘K-31’ while it did not change in GA-treated ‘Bonsai’. XET3 was not responsive to GA or TE treatment in ‘K-31’, but increased with GA treatment for ‘Bonsai’.
(A) EXPA4; (B) EXPA5; (C) EXPA7. The vertical bar is the standard error of mean (n = 4 replicates of GA or TE treatment, and each replicate contains at least 10 seedlings). Columns marked with different letters indicate significant differences between treatments and between cultivars based on LSD test (P = 0.05).
(A) EXPB4; (B) EXPB7. The vertical bar is the standard error of mean (n = 4 replicates of GA or TE treatment, and each replicate contains at least 10 seedlings). Columns marked with different letters indicate significant differences between treatments and between cultivars based on LSD test (P = 0.05).
(A) XET1; (B) XET2; (C) XET3. The vertical bar is the standard error of mean (n = 4 replicates of GA or TE treatment, and each replicate contains at least 10 seedlings). Columns marked with different letters indicate significant differences between treatments and between cultivars based on LSD test (P = 0.05).
Discussion
The leaf elongation rate and elongation duration time are two main factors contributing to the leaf growth. Here, the leaf elongation profiles of two tall fescue genotypes with contrasting elongation rates were examined. The leaf elongating duration times of ‘K-31’ and ‘Bonsai’ were similar (see Supplemental Fig. S2) and the leaf length differences between them were mostly due to the differences of leaf elongation rates. In a study of barley, Rht3 dwarfing gene led the plants to a shorter leaf length compared with rht3 wild type, along with reduced growth rate but same growth duration time39. Rht3 mutant is generally supposed to loss the response to gibberellin, which is an important phytohormone greatly promoting the leaf elongation in many species, such as wheat43,44, barley45, and maize46.
The elongation zone is a sensitive factor influenced by genetic variations and GA regulation. Jovanovic et al.47 found in maize that the length of the growth zone in high-LER cultivar was 25% longer than that in low-LER cultivar47. Another study in wheat also reported that a GA-deficient dwarf mutant, M489, had reduced leaf length and maximum REGR and further experiments documented that exogenous gibberellic acid increased the leaf length and REGR of mutant plants up to the level of wild type. The results above inferred GA regulation might attribute to the genetic variation in leaf growth through controlling the length of leaf elongation zone and REGR. In this study, fast-growing ‘K-31’ leaves had longer (44%) leaf elongation zone than slow-growing ‘Bonsai’, and the maximum REGR of ‘K-31’ was significantly higher than ‘Bonsai’. Further, the endogenous GA content of two genotypes were examined and results indicated that the variations in LER between two tall fescue cultivars were associated with differential levels of endogenous GA4, with fast-growing ‘K-31’ leaves having greater GA4 content than slow-growing ‘Bonsai’ leaves.
Exogenous application of GA3 further stimulated LER in both cultivars. Similar results were found in leaves of soybeans (Glycine max) treated with different concentrations of GA3, which demonstrated that the increase of plant height with GA treatment was positively associated with the increased level of endogenous GA48. In our study, LER of ‘K-31’ was higher than ‘Bonsai’ after GA3 application, which was corresponded with the endogenous GA1 content. In addition, elongation of both interstomatal cells and long cells were stimulated by exogenous GA treatment and inhibited by TE treatment in our study, which suggested that GA could play regulatory roles in leaf elongation through controlling the cell elongation in tall fescue.
Variations in leaf elongation rate could also be due to differential elongation rates of different cell types. Tina et al. (2005) reported that the interstomatal cells in leaf blade had the same length between semi-dwarf and tall cultivars of wheat while the long cells in tall cultivars were much longer than the semi-dwarf cultivar36. Hu and Schmidhalter49 found that the reduction in leaf elongation in wheat by salinity stress was due to the inhibition of the long cell growth while the length of interstomatal cells remained unchanged under salinity stress49. In our study, interstomatal cell length did not differ between fast-growing ‘K-31’ and slow-growing ‘Bonsai’ whereas the long cells in ‘K-31’ were significantly longer than ‘Bonsai’, suggesting that the genetic variation in LER in tall fescue was largely associated with the differences in the growth rate of long cells, which could be manipulated through genetic modification generating fast- or slow-growing phenotypes.
Despite the knowledge of genetic variations and the well-known simulative effect of GA on leaf growth rate, few reports provide the information about the relationship between the GA response and expansin or XET genes expression in relation to the genetic variations in leaf growth. In our study, the expression levels of EXPA7 were significantly higher in ‘K-31’ than ‘Bonsai’ with or without GA treatment, which inferred that the EXPA7 was attributed to the higher leaf elongation rate of ‘K-31’ with their relaxation functions in the cell wall. Other expansin genes and XET genes tested in this study did not show consistent differences in their expression levels between the fast-growing and slow-growing cultivars, which might because the expansins and XETs are both families of genes, different members may play various roles in plant growth and development processes50,51. For instance, in a study with Lycopersicon esculentum, the expression of LeEXP2 was positively associated with the elongation rate of hypocotyls, but the expression level of LeEXP18 did not correlate with hypocotyl growth rate52.
GA effects on leaf elongation and genetic variations in leaf elongation could be due to the involvement of different cell-wall loosening genes. In our study, among five homologous expansin genes, three α-expansins (EXPA4, EXPA5, and EXPA7) and one β-expansin (EXPB4) were up-regulated by GA treatment in both cultivars, suggesting that those expansin genes could play roles in GA-enhanced cell elongation in tall fescue. The expression level of EXPA5 was greatly up-regulated by GA treatment in both genotypes, suggesting that it could be induced by high GA level. In the high-LER genotype ‘K-31’, however, which had higher endogenous GA content than ‘Bonsai’, EXPA5 expression level was lower than ‘Bonsai’ both with and without GA treatment. One explanation about this might be the gene EXPA5 in the ‘K-31’ was not as sensitive to GA as it in ‘Bonsai’, due to less GA receptors or transcriptional factors.
Differential response patterns of XET genes were found between the two cultivars in response to GA, with XET1 and XET2 being up-regulated in ‘K-31’ and XET3 up-regulated in ‘Bonsai’ with GA treatment. The results indicated that members of gene family are likely to play different roles in various plant genotypes within the same species. The results above together suggested that EXPA4 and EXPA7 could be more important accounting for the genetic variations in leaf cell elongation in tall fescue, while EXPA4, EXPA5, EXPA7, EXPB4, XET1, XET2 and XET3 could be regulated by GA, although multiple cell-wall loosening genes could coordinately regulate cell elongation controlled genetically or regulated by GA.
Conclusions
Taken together, this study demonstrated that the genetic variation in leaf elongation of tall fescue cultivars with differential growth rate were associated with the differential leaf elongation zones differing in both cell elongation rate and production rate. Cultivar differences in endogenous GA content and exogenous treatment of plants with GA or TE suggested that GA could play roles in regulating leaf elongation in association with the up-regulation of several expansin and XET genes. Those cell-wall loosening genes, including EXPA4 and EXPA7, related to genetic variations and responsive to GA (EXPA4, EXPA5, EXPA7, EXPB4, XET1, XET2 and XET3), could be used as potential candidate genes to modify genetically grass leaves for rapid leaf elongation as needed in forage grass or slow leaf elongation as a desirable trait for turf grass through gene over-expression or knockout.
Materials and Methods
Plant materials and growth conditions
Seedlings of tall fescue (‘K-31’ and ‘Bonsai’) were established from seeds planted in plastic containers filled with fritted clay. Seedlings were watered daily and fertilized weekly with half-strength Hoagland’s nutrient solution53. Plants were maintained in a walk-in growth chamber (Environmental Growth Chambers, Chagrin Falls, OH) controlled at 22/18 °C (day/night) temperature, 60% relative humidity, and 12-h photoperiod with photosynthetically-active radiation of 750 μmol m−2s−1 at the canopy level. The experiments were conducted at Rutgers University between March and June in 2015. Grasses for the genetic variation analysis of LER and REGR were seeded on 15th, March, 2015 and the first leaf emerged mostly on 21th, March, which was recorded as the Day 1 of first leaf elongation. Most of second leaves appeared on 28th, March, which was recorded as the Day 1 of second leaf elongation, while the Day 1 of third leaf was 4th, April.
Gibberellin (GA) and gibberellin inhibitor treatments
Grasses for GA regulation experiment were seeded on 1st, April, 2015 and the for the uniformity of GA treatments, a hydroponic system was used in a growth chamber as previous described54. Uniform size one-week old seedlings at 1.2 (Haun Index) leaf stage55 established under the above conditions were transferred (14th, April) to plastic containers (54 cm in length, 42 cm in width, and 14 cm in depth) containing modified Hoagland’s nutrient solution. The nutrient solution contained ammonium sulfate ((NH4)2SO4, 71.36 mg L−1), potassium nitrate (KNO3, 27.3 mg L−1), calcium nitrate tetrahydrate (Ca(NO3)2.4H2O, 120.8 mg L−1), potassium phosphate monobasic (KH2PO4, 81.65 mg L−1), potassium sulfate (K2SO4, 52.28 mg L−1), magnesium sulfate anhydrous (MgSO4, 60 mg L−1), EDTA, ferric sodium salt trihydrate (Fe(EDTA)Na, 16.84 mg L−1), boric acid (H3BO3, 1.43 mg L−1), manganese chloride (MnCl2.4H2O, 0.91 mg L−1), zinc sulfate (ZnSO4. H2O, 0.11 mg L−1), cupric sulfate (CuSO4, 0.04 mg L−1), ammonium molybdate ((NH4)Mo7O24.4H2O, 0.01 mg L−1). The nutrient solution was aerated by air pumps (115 V, 60 Hz, Tetra Blacksburg, VA) and changed every 5 d. The pH of nutrient solution was adjusted every other day to 5.8 using KOH.
For the investigation of GA regulation of leaf elongation, seedlings were treated with GA3 (Sigma-Aldrich, St. Louis, MO) or a GA inhibitor, trinexapac-ethyl (TE) on 24th, April. For GA3 treatment, roots of plants was immersed in the nutrient solution with GA3 for 12 h, and leaf elongation rate was evaluated for 3 days after GA treatment. Different concentrations (0, 10, 25, 50, 100, and 200 μmol L−1) of GA3 were tested and the 50 μmol L−1 was found to be most effective in promoting leaf elongation and was selected to use in the subsequent experiment. For TE, the product Primo Maxx (Syngenta Professional Products, Greensboro, NC) was sprayed onto the leaves of grasses until dripping at the manufacture recommended concentration (2 mL L−1 [v/v]; a.i. TE = 11.3%) for tall fescue plants. The untreated control plants were sprayed with equal volume of water as used in TE treatment.
The experiment with GA or TE treatments for two cultivars were arranged in a split-plot design with GA or TE treatment as main plots (4 containers for each treatment) and cultivars as sub-plots (40 plants for each cultivar) which were randomly placed within each container with or without GA or TE treatment. The GA or TE treatment was repeated in four containers as four replicates. Each cultivar had 40 plants as replicates for each sampling of leaves for various measurements described below. Each plant was wrapped at the base of the plant with a sponge strip which was placed in a hole of a styrofoam plate, holding the plant in upright position in each container.
Leaf elongation rate (LER) and relative elemental growth rate (REGR)
The length of the first three leaves of both plant cultivars (‘K-31’ and ‘Bonsai’) was monitored from the first day of the leaf emergence till the third leaves were fully expanded, and the leaf elongation rate (LER, mm d−1) for each leaf was calculated as the slope of the linear regression line through the data points within the phase of linear increase in leaf length. The linear growth phase of the leaves was determined as the interval between 20 and 80% of final leaf length38. If we described the function of leaf length (y) to days of leaf elongation (x) as equation (1) y = mx + b, the slope m was the LER of each leaf. The linear regression line was drawn by Excel with the data plots selected within the linear interval and the LER (m) was calculated by the equation (2) m = [n∑(xy) − ∑x∑y]/[n∑(x2) − (∑x)2], which x is the days of leaf elongation, y is the leaf length and n is the days of recorded. The R2 is the square of the correlation coefficient.
The length of the third leaf on each plant for each cultivar and GA3 or TE treatment was measured using a ruler every day, beginning when the leaf tip just emerged. The REGR was a series of segmental growth rates along elongating leaves and determined by the leaf length increment within 24 h56. The spatial distribution of leaf REGR was determined using a pinhole method10. As previous reports indicated, the spatial distribution of REGR was steady between the emergence of the leaf tip and the transition from blade to sheath growth10. Therefore, a set of 10 plants per replicate was selected when the third leaf shortly emerged. Holes were pinned with fine needles (0.25 mm diameter) through the sheaths of the outer leaves and the first hole is 2 mm from the leaf base. The elongation zone of tall fescue is restricted within the base region of the leaf, and 30 holes spaced 2 mm apart were pierced along the longitudinal axial of leaf. After 24 h, the third leaves were removed from the plant and observed under a stereomicroscope fitted with a camera (Nikon Instruments Inc., model SMZ1270, Melville, NY, USA). The final positions of the holes along the third leaves were recorded using the camera. The relative elemental growth rate (REGR, mm mm−1 h−1) was calculated as described by Arredondo12: REGR = (df − di)/(di × Δt) where df (mm) and di (mm) is the final and initial distances between two holes along the leaves and the Δt (h) is the time period between pinning and observation.
Epidermal cell length and cell production rate
The third leaves were harvested when they were fully expanded. Leaves were cut at the base of leaf blade and transferred into methanol immediately for chlorophyll removal. Then the leaves were transferred to 85% lactic acid for storage. The abaxial surface of leaf was brushed by nail polish and a fine transparent negative film of the epidermis was obtained. The picture of epidermis was observed under microscope and captured by a camera. The length of two kinds of epidermal cells (long cells and interstomatal cells) are measured by software Digimizer (MedCalc Software bvba, Ostend, Belgium) and compared among different treatments36. The characteristics of each type of epidermal cells were according to the previous report of Botwright36 in regard to the wheat leaf. The interstomatal cells are the subsidiary cells between two stomata and long cells are long, unspecialized cells between the two cell rows of interstomatal and guard cells (see Supplemental Fig. S1). Cell production rates of both kinds of epidermis cells were calculated based on the mature cell length (Lm) and leaf elongation rate using the equation P = LER (mm d−1) ×Lm (mm)−1, assuming during the steady-state leaf elongation, the flux of cells through any point in the elongation-only zone is constant and represents the rate of cell production (P, cells d−1)3. Epidermal cell length and cell division rate were determined on 40 leaves from 40 plants for each cultivar subjected to GA or TE treatment. For each leaf, 50 cells were measured and the average length and cell production rate was taken to use for the further analysis.
Quantification of GA content in leaves
Gibberellic acid extraction and quantification was based on the method used by57. Frozen tissue samples were lyophilized using VirTis Genesis freeze dryer (SP Scientific, model 12 EL, NY). Lyophilized samples was ground to a fine powder using Genogrinder 2000 (OPS Diagnostics, model SP2100-115, NJ) and approximately 50 mg was weighed and used for the hormone extraction and analysis. Extractions were handled in the same manner as described for kentucky bluegrass (Poa pratensis) in Krishnan and Merewitz (2014)58. 100 nmol of deuterium-labeled GA4 (d2-GA4) and GA1 (GA1) was added at the time of extraction as the internal standard for liquid chromatography (LC) analysis. GA content was analyzed using an Ultra High-performance Liquid Chromatography-tandem mass spectrometer (UPLC/MS/MS) (Waters Quattro Premier XE ACQUITY® Tandem Quadrupole, Waters, Milford, MA).
Gene expression analysis
The expression level of expansin and XET genes in leaves under different treatments were examined using qPCR (quantitative Polymerase Chain Reaction). The entire tall fescue EST (Expressed Sequence Tags) database in NCBI (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/) was searched using expansin and XET gene families of rice, wheat, meadow fescue and Brachypodium distachyon based on the similarity, and 5 expansin ESTs and 3 XET ESTs were found in the tall fescue EST database. Primers of 5 expansin ESTs and 3 XET genes for quantitative PCR were designed by Primer3 using those sequences which are listed Table 1. Because meadow fescue and tall fescue have very similar genetic background, the gene PpXET3 of meadow fescue, which was not found to have a homologous sequence in tall fescue’s EST database, was used to design the qPCR primers along with the other EST sequences.
The third leaves of each plant were harvested and frozen in liquid nitrogen for the RNA extraction. Total RNA was extracted using Trizol reagent (Life Technologies, Grand Island, NY) and the contaminating DNA was removed by TURBO DNA-free kit (Life Technologies, Grand Island, NY). Reverse transcription of total RNA to single strand DNA was performed by the high capacity cDNA reverse transcription kit (Life Technologies, Grand Island, NY). The qPCR was performed in StepOne Real-time PCR System (Life Technologies, Grand Island, NY) and the cycling condition was 95 °C for 10 mins, 40 cycles of 95 °C denaturation for 30 seconds and 60 °C annealing/extension for 60 seconds, 95 °C for 30 seconds, followed by dissociation curve analysis. Power SYBR Green PCR Master Mix (Life Technologies, Grand Island, NY) was the intercalating dye used to detect gene expression level. Gene name, accession number, forward and reverse primer sequences are provided in Table 1. A tall fescue actin gene was used as the reference gene59 and a ΔΔCt method was used to calculate the relative expression level of interest and reference genes.
Statistical analysis
All data were subjected to the analysis of variance test using the general linear model with a statistical program (SAS 9.0, Cary, NC). The differences between GA, TE and the untreated control treatments and cultivar variations in LER, cell elongation rate, cell production rate, and gene expression levels were also tested using the least significance test at probability level of 0.05.
Additional Information
How to cite this article: Xu, Q. et al. Gibberellin-Regulation and Genetic Variations in Leaf Elongation for Tall Fescue in Association with Differential Gene Expression Controlling Cell Expansion. Sci. Rep. 6, 30258; doi: 10.1038/srep30258 (2016).
References
Horst, G. L., Nelson, C. J. & Asay, K. H. Relationship of leaf elongation to forage yield of tall fescue genotype. Crop Science 18, 715–719 (1978).
Brosse, N., Dufour, A., Meng, X., Sun, Q. & Ragauskas, A. Miscanthus: a fast‐growing crop for biofuels and chemicals production. Biofuels, Bioproducts and Biorefining 6, 580–598 (2012).
Bultynck, L., Fiorani, F., Van, V. E. & Lambers, H. Epidermal cell division and cell elongation in two Aegilops species with contrasting leaf elongation rates. Functional plant biology 30, 425–432 (2003).
Skinner, R. & Nelson, C. Elongation of the grass leaf and its relationship to the phyllochron. Crop Science 35, 4–10 (1995).
MacAdam, J. W., Volenec, J. J. & Nelson, C.J. Effects of nitrogen on mesophyll cell division and epidermal cell elongation in tall fescue leaf blades. Plant Physiology 89, 549–556 (1989).
Durand, J. L., Onillon, B., Schnyder, H. & Rademacher, I. Drought effects on cellular and spatial parameters of leaf growth in tall fescue. Journal of experimental botany 46, 1147–1155 (1995).
Cramer, G. R. Differential effects of salinity on leaf elongation kinetics of three grass species. Plant and Soil 253, 233–244 (2003).
Fagerness, M. J., Isgrigg, J., Cooper, R. J. & Yelverton, F. H. Plant growth regulators and mowing height affect ball roll and quality of creeping bentgrass putting greens. HortScience 35, 755–759 (2000).
Salas Fernandez, G. M., Becraft, P. W., Yin, Y. & Lübberstedt, T. From dwarves to giants? Plant height manipulation for biomass yield. Trends in Plant Science 14, 454–461, doi: http://dx.doi.org/10.1016/j.tplants.2009.06.005 (2009).
Schnyder, H., Seo, S., Rademacher, I. & Kühbauch, W. Spatial distribution of growth rates and of epidermal cell lengths in the elongation zone during leaf development in Lolium perenne L. Planta 181, 423–431 (1990).
Skinner, R. & Nelson, C. Epidermal cell division and the coordination of leaf and tiller development. Annals of Botany 74, 9–16 (1994).
Arredondo, J. T. & Schnyder, H. Components of leaf elongation rate and their relationship to specific leaf area in contrasting grasses. New Phytologist 158, 305–314 (2003).
Fiorani, F., Beemster, G. T., Bultynck, L. & Lambers, H. Can meristematic activity determine variation in leaf size and elongation rate among four Poa species? A kinematic study. Plant Physiology 124, 845–856 (2000).
Volenec, J. J. & Nelson, C. J. Cell dynamics in leaf meristems of contrasting tall fescue genotypes. Crop Science 21, 381–385 (1981).
Cosgrove, D. J. Loosening of plant cell walls by expansins. Nature 407, 321–326 (2000).
Ookawara, R., Satoh, S., Yoshioka, T. & Ishizawa, K. Expression of α-expansin and xyloglucan endotransglucosylase/hydrolase genes associated with shoot elongation enhanced by anoxia, ethylene and carbon dioxide in arrowhead (Sagittaria pygmaea Miq.) tubers. Annals of Botany 96, 693–702 (2005).
Cosgrove, D. J. Cell wall loosening by expansins. Plant Physiology 118, 333–339 (1998).
Kende, H. et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Molecular Biology 55, 311–314 (2004).
Sampedro, J. & Cosgrove, D. J. The expansin superfamily. Genome Biology 6, 242 (2005).
Choi, D., Cho, H. T. & Lee, Y. Expansins: expanding importance in plant growth and development. Physiologia plantarum 126, 511–518 (2006).
Muller, B. et al. Association of specific expansins with growth in maize leaves is maintained under environmental, genetic, and developmental sources of variation. Plant Physiology 143, 278–290 (2007).
Lee, Y. & Kende, H. Expression of α-expansin and expansin-like genes in deepwater rice. Plant Physiology 130, 1396–1405 (2002).
Li, F. et al. Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. Journal of plant physiology 168, 960–966 (2011).
Reidy, B., McQueen-Mason, S., Nösberger, J. & Fleming, A. Differential expression of α-and β-expansin genes in the elongating leaf of Festuca pratensis . Plant Molecular Biology 46, 491–504 (2001).
Vriezen, W. H., De Graaf, B., Mariani, C. & Voesenek, L. A. C. J. Submergence induces expansin gene expression in flooding-tolerant Rumex palustris and not in flooding-intolerant R. acetosa. Planta 210, 956–963, 10.1007/s004250050703 (2000).
Azeez, A., Sane, A. P., Tripathi, S. K., Bhatnagar, D. & Nath, P. The gladiolus GgEXPA1 is a GA-responsive alpha-expansin gene expressed ubiquitously during expansion of all floral tissues and leaves but repressed during organ senescence. Postharvest Biology and Technology 58, 48–56 (2010).
Lü, P. et al. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 237, 1547–1559 (2013).
Goh, H.-H., Sloan, J., Carmen, D. F. & Fleming, A. Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiology 159, 1759–1770 (2012).
Choi, D., Lee, Y., Cho, H. T. & Kende, H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. The Plant Cell 15, 1386–1398 (2003).
Eklöf, J. M. & Brumer, H. The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiology 153, 456–466 (2010).
Smith, R. C., Matthews, P. R., Schünmnn, P. H. & Chandler, P. M. The regulation of leaf elongation and xyloglucan endotransglycosylase by gibberellin in ‘Himalaya’barley (Hordeum vulgare L.). Journal of experimental botany 47, 1395–1404 (1996).
Palmer, S. J. & Davies, W. J. An analysis of relative elemental growth rate, epidermal cell size and xyloglucan endotransglycosylase activity through the growing zone of ageing maize leaves. Journal of experimental botany 47, 339–347 (1996).
Achard, P. et al. Gibberellin Signaling Controls Cell Proliferation Rate in Arabidopsis. Current Biology 19, 1188–1193, doi: http://dx.doi.org/10.1016/j.cub.2009.05.059 (2009).
Bolduc, N. & Hake, S. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. The Plant Cell 21, 1647–1658 (2009).
Luo, A. et al. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant and cell physiology 47, 181–191 (2006).
Botwright, T. L., Rebetzke, G. J., Condon, A. G. & Richards, R. A. Influence of the gibberellin-sensitive Rht8 dwarfing gene on leaf epidermal cell dimensions and early vigour in wheat (Triticum aestivum L.). Annals of Botany 95, 631–639 (2005).
Li, J. et al. Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. The Plant Cell 23, 628–640 (2011).
Bultynck, L. & Lambers, H. Effects of applied gibberellic acid and paclobutrazol on leaf expansion and biomass allocation in two Aegilops species with contrasting leaf elongation rates. Physiologia plantarum 122, 143–151 (2004).
Tonkinson, C. L., Lyndon, R. F., Arnold, G. M. & Lenton, J. R. Effect of the Rht3 dwarfing gene on dynamics of cell extension in wheat leaves, and its modification by gibberellic acid and paclobutrazol. Journal of experimental botany 46, 1085–1092 (1995).
Wenzel, C. L., Williamson, R. E. & Wasteneys, G. O. Gibberellin-induced changes in growth anisotropy precede gibberellin-dependent changes in cortical microtubule orientation in developing epidermal cells of barley leaves. Kinematic and cytological studies on a gibberellin-responsive dwarf mutant, M489. Plant Physiology 124, 813–822 (2000).
Huang, B. & Gao, H. Physiological responses of diverse tall fescue cultivars to drought stress. HortScience 34, 897–901 (1999).
Huang, B., Fry, J. & Wang, B. Water relations and canopy characteristics of tall fescue cultivars during and after drought stress. HortScience 33, 837–840 (1998).
Rebetzke, G. et al. Height reduction and agronomic performance for selected gibberellin-responsive dwarfing genes in bread wheat (Triticum aestivum L.). Field Crops Research 126, 87–96 (2012).
Zhang, Y., Ni, Z., Yao, Y., Nie, X. & Sun, Q. Gibberellins and heterosis of plant height in wheat (Triticum aestivum L.). BMC genetics 8, 40 (2007).
Asthir, B., Duffus, C. M. & Spoor, W. Correlation of gibberellin-induced growth, polyamine levels and amine oxidases in epicotyl, root and leaf blade of barley seedlings. Plant growth regulation 42, 193–201 (2004).
Nelissen, H. et al. A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Current Biology 22, 1183–1187 (2012).
Jovanovic, Z., Djakovic, T., Stikic, R., Prokic, L. & Sukalovic, V. H.-T. Effect of N deficiency on leaf growth and cell wall peroxidase activity in contrasting maize genotypes. Plant and soil 265, 211–223 (2004).
Hamayun, M. et al. Exogenous gibberellic acid reprograms soybean to higher growth and salt stress tolerance. Journal of Agricultural and Food Chemistry 58, 7226–7232 (2010).
Hu, Y. C. & Schmidhalter, U. Effect of salinity on the composition, number and size of epidermal cells along the mature blade of wheat leaves. Journal of Integrative Plant Biology 49, 1016–1023 (2007).
Jung, J., O’Donoghue, E. M., Dijkwel, P. P. & Brummell, D. A. Expression of multiple expansin genes is associated with cell expansion in potato organs. Plant Science 179, 77–85 (2010).
Lizana, X. C. & Calderini, D. F. Expansins expression is associated with grain size dynamics in wheat (Triticum aestivum L.). Journal of experimental botany 61, 1147–1157 (2010).
Caderas, D. et al. Limited correlation between expansin gene expression and elongation growth rate. Plant Physiology 123, 1399–1414 (2000).
Arnon, D. I. Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiology 24, 1–15 (1949).
Merewitz, E. B., Gianfagna, T. & Huang, B. Photosynthesis, water use, and root viability under water stress as affected by expression of SAG12-ipt controlling cytokinin synthesis in Agrostis stolonifera. Journal of experimental botany 62, 383–395, 10.1093/jxb/erq285 (2011).
Haun, J. R. Visual quantification of wheat development. Agronomy Journal 65, 116–119, 10.2134/agronj1973.00021962006500010035x (1973).
Peters, W. S. & Bernstein, N. The determination of relative elemental growth rate profiles from segmental growth rates (a methodological evaluation). Plant Physiology 113, 1395–1404 (1997).
Liu, H., Li, X., Xiao, J. & Wang, S. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant methods 8, 1–12 (2012).
Krishnan, S. & Merewitz, E. B. Drought Stress and Trinexapac-ethyl Modify Phytohormone Content Within Kentucky Bluegrass Leaves. J Plant Growth Regul 34, 1–12 (2015).
Lü, S., Fan, Y. & Jin, C. Overexpression of a Ran GTPase homologous gene, FaRan from tall fescue, in transgenic Arabidopsis . Biologia Plantarum 55, 331–334 (2011).
Author information
Authors and Affiliations
Contributions
Designed the studies: B.H. Undertook the experimental work: Q.X. and S.K. Analyzed the data: Q.X and B.H. Contributed to figure and manuscript preparation: Q.X., B.H., E.M. and J.X.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Xu, Q., Krishnan, S., Merewitz, E. et al. Gibberellin-Regulation and Genetic Variations in Leaf Elongation for Tall Fescue in Association with Differential Gene Expression Controlling Cell Expansion. Sci Rep 6, 30258 (2016). https://doi.org/10.1038/srep30258
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep30258
This article is cited by
-
Split application of gibberellins at different growth stages to improve the yield and quality of cape gooseberry (Physalis peruviana L.) under subtropical plain
Vegetos (2024)
-
A protocol for in vitro propagation of Morella pubescens: a protected species in the Tambillo community protected area—Ecuador
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
Comparing the structural characteristics and expression of GA2ox gene in dwarf banana and its wild type
Journal of Plant Biochemistry and Biotechnology (2023)
-
Gibberellin-Induced Alterations to the Expression of Cell Wall-Related Genes in the Xylem of Carrot Root
Journal of Plant Growth Regulation (2021)
-
Stomatal response to drought is modulated by gibberellin in tomato
Acta Physiologiae Plantarum (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.