Abstract
Inferring the internal interaction patterns of a complex dynamical system is a challenging problem. Traditional methods often rely on examining the correlations among the dynamical units. However, in systems such as transcription networks, one unit's variable is also correlated with the rate of change of another unit's variable. Inspired by this, we introduce the concept of derivative-variable correlation and use it to design a new method of reconstructing complex systems (networks) from dynamical time series. Using a tunable observable as a parameter, the reconstruction of any system with known interaction functions is formulated via a simple matrix equation. We suggest a procedure aimed at optimizing the reconstruction from the time series of length comparable to the characteristic dynamical time scale. Our method also provides a reliable precision estimate. We illustrate the method's implementation via elementary dynamical models and demonstrate its robustness to both model error and observation error.
Similar content being viewed by others
Introduction
Modern world relies heavily on complex interconnected systems, such as Internet and social media or transport and communication networks. In addition to technological utilities, complex systems are found on various scales in nature and society: gene regulation, protein and metabolic networks, food webs, economic and social networks, climate. Among the foremost problems here is the development of methods for reconstructing the structure (topology) of real networks from the observable data. Topology, in combination with the inter-unit interactions, determine the function of complex dynamical systems (networks)1. Reconstruction methods are often developed within the contexts of particular fields, relying on domain-specific approaches. These include gene regulations2,3,4,5, metabolic systems6, neuroscience7, or social networks8. On the other hand, theoretical reconstruction concepts are based on paradigmatic dynamical models such as phase oscillators9,10,11,12, some of which have been experimentally tested13,14. In a similar context, techniques for detecting hidden nodes in networks are being investigated15. A class of general reconstruction methods exploit the time series obtained by quantifying the system behaviour. Some of them assume the knowledge of the internal interaction functions16,17, while others do not18. Network couplings can be examined via an information-theoretic approach19. The advantage of these methods is that they are non-invasive, i.e. require no interfering with the on-going dynamics.
Reconstruction methods are often based on examining the dynamical correlations among the system units (network nodes)12. On the other hand, models of complex dynamical systems such as Eq.1, are usually based on expressing the time derivative of a node as a combination of a local and a coupling term. Inspired by this, we propose a non-invasive reconstruction method, relying on the concept of derivative-variable correlation. Our method assumes the dynamical time series to be available as measured observables and the interaction functions to be known. We present our theory in a general form, extending our initial results20. As we show, our approach allows for the reconstruction precision to be estimated, indicating the level of noise in the data, or possible mismatches in the knowledge of the interaction functions.
Results
The reconstruction method
We consider a general complex network consisting of N nodes, described by their dynamical states xi(t). Its time evolution is governed by:

where the function fi represents the local dynamics for each node and hj models the action of the node j on other nodes. The network topology is encoded in the adjacency matrix Aji, specifying the strength with which the node j acts on the node i. We assume that: (i) the complex system evolves according to Eq.1, (ii) the interaction functions fi and hj are precisely known and (iii) a discrete trajectory consisting of L values xi(t1), …, xi(tL) is known for each node. The measurements of xi are separated by the uniform observation interval δt defining the time series resolution. We seek to reconstruct the unknown adjacency matrix Aij ≡ A under these assumptions.
The starting point is to define the following correlation matrices, using the observable g(x) whose role will be explained later:

where 〈·〉 denotes time-averaging  . Inserting into the Eq.1, we obtain the following linear relation between the correlation matrices:
. Inserting into the Eq.1, we obtain the following linear relation between the correlation matrices:

which is our main reconstruction equation, applicable to any system with dynamics given by Eq.1. Time series are to be understood as the available observables, allowing for matrices in Eq.2 to be computed for any g. For the infinitely long dynamical data, reconstruction is always correct for any non-trivial choice of g. For short time series, representing experimentally realistic scenarios, the reconstruction is always approximate and its precision crucially depends on the choice of g (usually, correlations are defined as central moments with averages subtracted – instead, we are here not interested in correlations per se, but in the reconstruction according to Eq.3, for which the subtraction of averages is not needed). To be able to quantify the reconstruction precision, we need to equip ourselves with the adequate measures. To differentiate from the original adjacency matrix A, we call the reconstructed matrix Rij ≡ R and express the matrix error as:

Of course, each R is computed according to Eq.3 in correspondence with the chosen g. However, since the matrix A is unknown, we have to introduce another precision measure, based only on the available data (time series and interaction functions). A natural test for each R is to quantify how well does it reproduce the original data xi(tm). We apply the following procedure: start the dynamics from xi(t1) and run it using R until t = t2; denote thus obtained values yi(t2); re-start the run from xi(t2) and run until t = t3, accordingly obtaining yi(t3) and so on. The discrepancy between the reconstructed time series yi(tm) and the original xi(tm) is an explicit measure of the reconstruction precision, based solely on the available data. We name it trajectory error ΔT and define it as follows:

Different choices of the observable g lead to different R, with different precisions expressed through errors ΔT and ΔA. As we show below, these two error measures are related, meaning that small ΔT suggests small ΔA. The function g hence plays the role of a tunable parameter, which can be used to optimize the reconstruction. By considering many R-s obtained through varying g, we can single out R-s with the minimal ΔT to obtain the best reconstruction.
Implementation of the method
To illustrate the implementation of our method, we begin by formulating a complex dynamical system as a network with N = 6 nodes. 17 directed links, modelling inter-node interactions, are put between randomly chosen pairs of nodes. We examine two different examples of interactions. As our first example, we consider the Hansel-Sompolinsky model, describing the dynamics of firing in neural populations21. It is defined by the interaction functions fi = −x and hj = tanh x which are fixed for all nodes. The adjacency matrix is specified by assigning positive and negative weights to the links, randomly chosen from [−10, 10], as shown in Fig. 1a. Weights model the varying strengths of interaction. Starting from random initial conditions, the resulting system is integrated from t = 0 to t = 4. During the run, 20 values of xi are stored for each node, equally spaced with δt = 0.2. The obtained time series, shown in Fig. 2, are rather short compared to the characteristic time scale and the system size.
We now use these data to reconstruct the original adjacency matrix by employing the procedure described above. We consider a set of 104 test-functions g, each composed of first 10 Fourier harmonics

The coefficients ak and bk are randomly selected from [0, 100] with the log-uniform probability (to emphasize smaller coefficient values). This is implemented by selecting each Fourier coefficient via 102.00432137×rand − 1.0, where rand is a random number between 0 and 1. A typical function thus constructed for each choice of ak and bk will have all 10 Fourier components, but one (or at most few) will be well pronounced. Functions are then normalized to the range of time series values. Given relatively smooth time series, lower harmonics are expected to generally extract more features from data, which is why we limit ourselves to the first 10 harmonics. To improve the stability of the derivative estimates, we base our calculations on the set of time points τm = (tm+1 − tm)/2. For each g, the matrix R is obtained via Eq.2 and Eq.3, with the invertibility of each E checked by virtue of the singular value decomposition. The errors ΔT and ΔA are then calculated for each R and reported as a scatter plot in Fig. 3a.
The main result of this analysis is a clear correlation between ΔT and ΔA, particularly pronounced for smaller values of errors. This confirms that the best R are among those that display minimal ΔT. In order to identify the best reconstruction and estimate its precision, we focus on the 1% of matrices R with the minimal ΔT, as illustrated in Fig. 3a by the dashed vertical line. The variability of R within this group can be viewed as the reconstruction precision. Small variability indicates the invariance of R to the choice of g, which suggests a good reconstruction. Large variability of R implies its drastic dependence on g, indicating a bad precision. We quantify this by computing the mean and the standard deviation for each matrix element of R within this group and identify them, respectively, with the best reconstruction value and its precision. In Fig. 4a we report the original A and the best reconstruction, along with the respective errorbar for each matrix element, describing the reconstruction precision. The reconstruction is indeed very good for both zero and non-zero weights (i.e. for non-linked and linked node pairs).
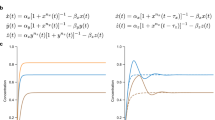
Our second example of node dynamics is a model describing gene interactions, with the coupling functions of two types: activation  and repression
and repression  22. Local interaction are again modeled via fi = −x. The adjacency matrix is based on the same network and defined by assigning a random weight from [0, 1] for each link, as shown in Fig. 1b. The nodes 1–3 (respectively, 4–6) act activatorily (repressively) on all nodes that they act upon. Again, we run the dynamics from t = 0 to t = 4, obtaining another set of time series with 20 points (not shown). The same reconstruction procedure is applied, yielding the ΔT vs. ΔA scatter plot shown in Fig. 3b. Using the same procedure, we obtain the best reconstruction and show it in Fig. 4b. The precision is again very good, although the relationship between ΔT and ΔA is now different, since the two examined systems display different degrees of non-linearity. This shows that our method applies even in cases of strongly non-linear interaction functions, which capture most real dynamical scenarios.
22. Local interaction are again modeled via fi = −x. The adjacency matrix is based on the same network and defined by assigning a random weight from [0, 1] for each link, as shown in Fig. 1b. The nodes 1–3 (respectively, 4–6) act activatorily (repressively) on all nodes that they act upon. Again, we run the dynamics from t = 0 to t = 4, obtaining another set of time series with 20 points (not shown). The same reconstruction procedure is applied, yielding the ΔT vs. ΔA scatter plot shown in Fig. 3b. Using the same procedure, we obtain the best reconstruction and show it in Fig. 4b. The precision is again very good, although the relationship between ΔT and ΔA is now different, since the two examined systems display different degrees of non-linearity. This shows that our method applies even in cases of strongly non-linear interaction functions, which capture most real dynamical scenarios.
Testing the method's robustness
In order to model the real applicability of our method, we test its robustness to possible violations of the initial assumptions, focusing on the first dynamical example (Fig. 1a). We start with the scenario when the interaction functions are not precisely known – we assume a small mismatch in their mathematical form (model error). Instead of the original fi = −x and hj = tanh x, we take fi = −1.1x and hj = tanh(1.1x) + 0.1x. The measurements of ΔT now cannot be expected to converge to zero. Nevertheless, we apply the same procedure and find (a weaker) correlation between ΔT and ΔA, as shown (by black dots) in Fig. 5a. To see the worsening of the precision clearly, grey dots show the original non-perturbed scatter plot from Fig. 3a. Dashed vertical line shows the part of the error ΔT which is unavoidable due to the presence of the perturbation. We compute it by averaging the difference between the original and the perturbed interaction functions over the y-range of the time series (for the function h we average  over the y-range of the time series and equivalently for the function f; since the two contributions can not be simply added, we consider only the larger one, in this case h, which gives the lower bound on such error). Such error is not related to the performance of our method, so we show it in the figure in order to isolate more clearly the part of the error that in fact arises from our method. The remaining ΔT is similar to the ΔT occurring in the non-perturbed case. This demonstrates that our method works even under perturbed conditions. The worsening of the reconstruction precision is what expected from the nature of the perturbation, meaning that our method makes no additional “unexpected” errors in the perturbed conditions. The best reconstruction and the corresponding errorbars are computed as before and shown in Fig. 6a. The errorbars are larger and the reconstruction precision worsens. Still, the essential fraction of elements of A are within the respective errorbars. The decline of precision is controllable, since it is clearly signalized by the size of the errorbars. This could be used to generalize the method in the direction of detecting the interaction functions as well. Each best R would be accompanied by the best guesses for fi and hj, meaning that different network topologies, reproducing the data equally well, would come in combination with different fi and hj.
over the y-range of the time series and equivalently for the function f; since the two contributions can not be simply added, we consider only the larger one, in this case h, which gives the lower bound on such error). Such error is not related to the performance of our method, so we show it in the figure in order to isolate more clearly the part of the error that in fact arises from our method. The remaining ΔT is similar to the ΔT occurring in the non-perturbed case. This demonstrates that our method works even under perturbed conditions. The worsening of the reconstruction precision is what expected from the nature of the perturbation, meaning that our method makes no additional “unexpected” errors in the perturbed conditions. The best reconstruction and the corresponding errorbars are computed as before and shown in Fig. 6a. The errorbars are larger and the reconstruction precision worsens. Still, the essential fraction of elements of A are within the respective errorbars. The decline of precision is controllable, since it is clearly signalized by the size of the errorbars. This could be used to generalize the method in the direction of detecting the interaction functions as well. Each best R would be accompanied by the best guesses for fi and hj, meaning that different network topologies, reproducing the data equally well, would come in combination with different fi and hj.
Scatter plots of ΔT vs. ΔA for model and observation error scenarios.
Scatter plots of errors ΔT and ΔA (black dots), for the model error scenario in (a) and the observation error scenario in (b). Original non-perturbed scatter plot from Fig. 3a is shown in gray for comparison. Vertical dashed lines depicts the part of the ΔT error which is unavoidable in the presence of the perturbation (see text).
To test the third assumption of our theory, we take the time series to be not precisely known due to observation errors. Uncorrelated white noise of intensity η = 0.4 is added, perturbing each value of the time series. Instead of the original data, we now consider one realization of the noisy data, as illustrated in Fig. 2 (interaction functions are the original ones). The central problem now is the computation of the derivatives, which are extremely sensitive to the noise. We employ the Savitzky-Golay smoothing filter23 as a standard technique of data de-noising, which allows for a good derivative estimation. Since the time series are short, we apply the smallest smoothing parameters (polynomial degree 2, window size 2). The reconstruction procedure is applied as before, using smoothed derivatives to compute matrix B in Eq.2. The scatter plot of ΔT vs ΔA is shown in Fig. 5b, again compared with the original plot and with the perturbation-induced unavoidable error indicated by the vertical line (computed as before, this time averaging the perturbation value over the y-range of the time series). The worsening of the precision is of a similar magnitude as in the model error scenario. The best reconstruction and the corresponding errorbars are reported in Fig. 6b. Note that the precision is again correctly captured by the size of the errorbars. In two cases from Fig. 6, the precision does not decline uniformly for all links. The analysis above shows that our reconstruction method is reasonably robust to both model and observation error. We found this robustness to be qualitatively independent of the realization of both these errors.
Discussion and Conclusions
We presented a method of reconstructing the topology of a general complex dynamical system from the time series with known interaction functions. Through conceptually novel approach, our method is formulated as an inverse problem using linear systems formalism24. Rather than relying on the correlations between the observed variables, it is based on the correlations between the variables and their time derivatives. Our method involves two important factors: it applies to the data that is relatively short, i.e. of the length comparable to the system size and to the characteristic time scale; and, it yields the errorbars as a by-product, correctly reflecting the reconstruction precision.
On the other hand, our theory relies on knowing (at least approximately) both the dynamical model Eq.1 and the interaction functions. While these assumptions might limit the immediate applicability of our method, our idea presents a conceptual novelty, potentially leading towards a more general and applicable reconstruction method. For example, we expect applicability in studies of interacting neurons in slices or cultures, where the properties of the individual neurons (i.e. functions f and h) can be relatively well established, while the adjacency matrix is unknown. In contrast, the application to problems such as brain fMRI activity patterns, where even the existence of a dynamical model like Eq.1 is questionable, appears at present not possible.
Our theory includes choosing the tunable observables g, which allow for the reconstruction to be optimized within the constrains of any given data. The question of constructing the optimal g which extracts the maximal extractable information, remains open. Our algorithm can be reiterated: once the 1% of the best R-s are found, one can examine the functions g leading to those 1% and repeat the procedure, sampling only the neighboring portion of the functional space. Alternatively, various evolutionary optimization algorithms could be used25. An important factor for the method's applicability is the dynamical regime behind the time series, which could be regular and stable (for example periodic) or chaotic and unstable (starting from general initial conditions, the transients are typically irregular). The former case is less reconstructible, because of a poor coverage of the phase space. In particular, synchronized dynamics, being essentially non-sensitive to the variations of the coupling coefficients, offers very little insight into the structure of the underlying system. Increasing the time series length is obviously of no help20. In contrast, the latter case contains more extractable system information and is potentially more reconstructible. Chaotic dynamics provides a better coverage of the phase space, adding new information with increasing length of the time series. Another issue is the applicability to large networks  and in particular, the dependence of precision on relationship between N and L. This relates to the possibility of quantifying the network information content of the available data.
and in particular, the dependence of precision on relationship between N and L. This relates to the possibility of quantifying the network information content of the available data.
Another limitation of our theory comes from the form of Eq.1. A similar theory could be developed for alternative scenarios, such as h specified by both source and target nodes. The real challenge here are the networks with non-additive inter-node coupling (i.e., the dynamical contribution to the node i is not a mere sum of neighbours' inputs). The key practical problem is that the mathematical forms of f and h are not (precisely) known for many real networks, although for certain systems they can be inferred with a reasonable confidence4,5. Noise always hinders the reconstruction, specially via derivative estimates. However, longer time series not only bring more information, but also allow for a better usage of smoothing. Finally, we note that the network reconstruction problem is “opposite” of the network design problem. Our method could be employed to design a system that displays given dynamics. However, while any system with  solves the design problem, in the reconstruction theory this creates the permanent issue of isolating the true system among those that exhibit
solves the design problem, in the reconstruction theory this creates the permanent issue of isolating the true system among those that exhibit  .
.
Finally, we note that the comparison among the methods aimed at inferring the internal structure of dynamical systems is of great interest. Besides providing a consistent evaluation of the performance of various methods, such comparison could help one choose the most suitable method for a given problem. However, the initial hypotheses for various methods are very diverse, which makes the objective comparison among them very hard, at least at the moment. Directly applicable methods3,18 rely on experimentally realistic assumptions, but often perform poorly. Theoretical reconstruction ideas (such as the present or9,10,11,12) are based on stronger assumptions and show better performance, although their concrete applicability is for now limited. Nevertheless, this remains an important and interesting avenue for future work.
Change history
25 June 2014
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
Barrat, A., Barthélemy, M. & Vespignani, A. Dynamical Processes on Complex Networks (Cambridge University Press, Cambridge, 2008).
Hecker, M. et al. Gene regulatory network inference: Data integration in dynamic models - A review. Biosystems 96, 86–103 (2009).
Emmert-Streib, F. et al. Statistical inference and reverse engineering of gene regulatory networks from observational expression data. Front. Genet. 3, 8 (2012).
Nelson, D. et al. Oscillations in NF-κB Signaling Control the Dynamics of Gene Expression. Science 306, 704–708 (2004).
Pigolotti, S., Krishna, S. & Jensen, M. Oscillation patterns in negative feedback loops. Proc. Nat. Acad. Sci. USA 104, 6533–6537 (2007).
Herrgård, M. J. et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nature Biotech. 26, 1155–1160 (2008).
Bullmore, E. & Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosc. 10, 186–198 (2009).
Eagle, N., Pentland, A. & Lazer, D. Inferring friendship network structure by using mobile phone data. Proc. Nat. Acad. Sci. USA 106, 15274 (2009).
Levnajić, Z. & Pikovsky, A. Network Reconstruction from Random Phase Resetting. Phys. Rev. Lett. 107, 034101 (2011).
Kralemann, B., Pikovsky, A. & Rosenblum, M. Reconstructing phase dynamics of oscillator networks. CHAOS 21, 025104 (2011).
Prignano, L. & Díaz-Guilera, A. Extracting topological features from dynamical measures in networks of Kuramoto oscillators. Phys. Rev. E 85, 036112 (2012).
Ren, J. et al. Noise Bridges Dynamical Correlation and Topology in Coupled Oscillator Networks. Phys. Rev. Lett. 104, 058701 (2010).
Blaha, K. et al. Reconstruction of two-dimensional phase dynamics from experiments on coupled oscillators. Phys. Rev. E 84, 046201 (2011).
Stankovski, T. et al. Inference of Time-Evolving Coupled Dynamical Systems in the Presence of Noise. Phys. Rev. Lett. 109, 024101 (2012).
Su, R., Wang, W. & Lai, Y. Detecting hidden nodes in complex networks from time series. Phys. Rev. E 85, 065201(R) (2012).
Shandilya, S. G. & Timme, M. Inferring network topology from complex dynamics. New J. Phys. 13, 013004 (2011).
Džeroski, S. & Todorovski, L. Equation discovery for systems biology: finding the structure and dynamics of biological networks from time course data. Curr. Opin. Biotech. 19, 360–368 (2008).
Hempel, S. et al. Inner Composition Alignment for Inferring Directed Networks from Short Time Series. Phys. Rev. Lett. 107, 054101 (2011).
Pompe, B. & Runge, J. Momentary information transfer as a coupling measure of time series. Phys. Rev. E 83, 051122 (2011).
Levnajić, Z. Derivative-variable correlation reveals the structure of dynamical networks. Eur. Phys. J. B 86, 298 (2013).
Hansel, D. & Sompolinsky, H. Solvable Model of Spatiotemporal Chaos. Phys. Rev. Lett. 71, 2710 (1993).
Widder, S., Schicho, J. & Schuster, P. Dynamic patterns of gene regulation 1: Simple two gene systems. J. Theor. Biol. 246, 395–419 (2007).
Simonoff, J. S. Smoothing Methods in Statistics (Springer, New York, 1996).
Hespanha, J. P. Linear Systems Theory (Princeton University Press, Princeton, 2009).
Levnajić, Z. Evolutionary design of non-frustrated networks of phase-repulsive oscillators. Sci. Rep. 2, 967 (2012).
Acknowledgements
Work supported by Creative Core FISNM-3330-13-500033 “Simulations” funded by the European Union, The European Regional Development Fund. The operation is carried out within the framework of the Operational Programme for Strengthening Regional Development Potentials for the period 2007–2013, Development Priority 1: Competitiveness and research excellence, Priority Guideline 1.1: Improving the competitive skills and research excellence. Work also supported by the DFG via project FOR868, by ARRS via program P1-0383 “Complex Networks”, project J1-5454 “Unravelling Biological Networks” and project L7-4119 “Coauthorship networks of slovenian scholars: Theoretical analysis and visualization user interface development”. We thank colleagues Michael Rosenblum and Mogens H. Jensen for useful discussions.
Author information
Authors and Affiliations
Contributions
AP contributed the initial idea, ZL developed and conducted numerical simulations. ZL wrote the manuscript, while ZL and AP reviewed it.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Levnajić, Z., Pikovsky, A. Untangling complex dynamical systems via derivative-variable correlations. Sci Rep 4, 5030 (2014). https://doi.org/10.1038/srep05030
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05030
This article is cited by
-
The ill-posedness of derivative interpolation and regularized derivative interpolation for band-limited functions by sampling
EURASIP Journal on Advances in Signal Processing (2020)
-
Reconstruction of noise-driven nonlinear networks from node outputs by using high-order correlations
Scientific Reports (2017)
-
Reconstruction of noise-driven nonlinear dynamic networks with some hidden nodes
Science China Physics, Mechanics & Astronomy (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.