Abstract
Hyaluronidases (HAases), particularly leech HAases, have attracted intense attention due to their broad applications in medical treatments and great potential for the enzymatic production of hyaluronan oligosaccharides. However, little is known about this third interesting family of HAases. Here, we applied the random amplification of cDNA ends polymerase chain reaction (RACE-PCR) approach to identify the first leech HAase-encoding gene. By combining protein engineering and high-density culture, we achieved high-level production (8.42 × 105 U ml−1) in the yeast Pichia pastoris secretory expression system. Compared with the commercial bovine testicular HAase, the recombinant leech HAase exhibited superior enzymatic properties. Furthermore, analysis of the hydrolytic process suggested that this novel enzyme adopts a nonprocessive endolytic mode, yielding a narrow-spectrum of specific HA oligosaccharides with different incubation times. Large-scale production of this novel leech HAase will not only greatly promote medical applications but also facilitate the enzymatic production of specific HA oligosaccharides.
Similar content being viewed by others
Introduction
Hyaluronic acid (HA) is a linear and unbranched high-molecular weight polysaccharide composed of repeating disaccharide D-glucuronic acid (GlcUA) and N-acetyl-D-glucosamine (GlcNAc) units linked through β-1,4 bonds (Supplementary Fig. S1). High-molecular-weight HA is widely distributed among various host tissues and participates in numerous physiological processes1 and the biological functions and applications of HA depend on its molecular mass2,3. In particular, low-molecular-weight HA oligosaccharides have unique biological activities3. Smaller HA oligosaccharides can stimulate fibroblast proliferation and collagen synthesis4 and can selectively kill many types of cancer cells via disruption of the receptor-hyaluronan interaction5,6,7. HA octasaccharides (HA8) and decasaccharides (HA10) have significant suppressive effects on cancer cells8 and HA tetrasaccharides (HA4) and hexasaccharides (HA6) can induce dendritic cell maturation via Toll-like receptor (TLR)-4 associated with the antigen-presenting cells of the innate immune system9. In addition, lower-molecular-weight HA oligosaccharides are easily absorbed by the body and serve as precursors for the synthesis of higher-molecular-weight HA molecules and other substances. Thus, a specific narrow spectrum of HA oligosaccharides could have broad applications in medicine, food and cosmetics. Low-molecular-weight HA is mainly produced by the degradation of high-molecular-weight HA by physical and chemical methods10. However, the products of these methods have a broad range of molecular weights (>3,000 Da), making it difficult to obtain HA oligosaccharides with specific molecular weights. Many chemical approaches have been developed for the de novo synthesis of HA oligosaccharides7. However, these complex processes are time-consuming, carbohydrate oligosaccharide backbones are rare and the substrate uridine diphosphate (UDP)-sugars are expensive, limiting the applications of these synthetic methods in large-scale production11. In contrast, the enzymatic production of HA oligosaccharides with a well-characterised hyaluronidase (HAase) is promising and attractive because of its unique advantages, such as mild operation conditions, high degradation rates and high product uniformity12.
HAases (also previously referred to as “spreading factors”) are a large class of glycosidases that predominantly degrade HA. Based on substrate specificities and hydrolysis products, HAases are commonly grouped into three families (Supplementary Fig. S1)13: hyaluronate lyases (EC 4.2.2.1, Streptococcus hyaluronate lyase), hyaluronate 4-glycanohydrolases (EC 3.2.1.35, Bovine testicular hyaluronidase, BTH) and hyaluronate 3-glycanohydrolases (EC 3.2.1.36, Leech HAase). Commercial BTH has been widely used in clinical medicine and its hydrolysis mechanism has been studied extensively14. The disadvantages of the enzymatic production of specific or narrow-spectrum HA oligosaccharides by BTH include the limited source material (bovine testes), its considerably high price and the broad range of degradation products15. Compared with BTH and Streptococcus hyaluronate lyase, leech HAase has higher substrate specificity and a narrow-spectrum of enzymatic products16,17. In addition, the use of recombinant leech HAase does not pose any risk of animal cross-infection. Therefore, high-level production of recombinant leech HAase would be of great significance for both clinical medical treatment (such as surgery, ophthalmology and internal medicine) and producing narrow-spectrum HA oligosaccharides at the industrial scale. However, no gene sequence has been identified and reported for the EC 3.2.1.36 group, which includes leech HAase.
In this work, we cloned and characterised the first leech HAase-encoding gene, LHyal. Through a combination of N-terminal engineering and fed-batch fermentation strategies, we achieved high-level production of leech HAases in recombinant Pichia pastoris. More importantly, by exploring the hydrolytic process and mechanism, we achieved enzymatic production of narrowband HA oligosaccharides. The construction of robust microbial factories for the large-scale production of recombinant HAase and the development of the enzymatic approach for the preparation of specific HA oligosaccharides would greatly promote progress in related research areas, such as the chemical synthesis of oligosaccharides and cancer therapy.
Results
In silico characterisation of a potential HAase from leech
After analysing the expressed sequence tag (EST) databases of Hirudo sp., a potential HAase gene (designated LHyal) (deposited under accession number KJ026763) encoding 489 amino acids (Fig. 1a) was amplified from leech cDNAs after three rounds of random amplification of cDNA ends polymerase chain reaction (RACE-PCR). Basic Local Alignment Search Tool (BLAST) analysis of the deduced amino acid sequence suggested that LHyal contains a putative conserved glycosyl hydrolase family 79 N-terminal domain. Further multiple sequence alignment (MSA) analysis using CLUSTALW demonstrated that LHyal has low sequence identity with other well-characterised HAases (Supplementary Fig. S2) and exhibits higher sequence identity (approximately 35%) with heparanases (EC 3.2.1.166) (Fig. 1a). The motif 168NIDWELGNEP177 is highly conserved, with the exception of the residue D, which is replaced by S in all heparanases. Interestingly, the central tetrapeptide 169IDWE172 motif, which contains the putative active site acidic residues D170 and E172, are conserved in all HAases (Supplementary Fig. S2). A neighbour-joining tree was constructed to uncover the phylogenetic relationship among the hyaluronidases and heparanases (Fig. 1d). LHyal is close to the clades of heparanases and β-glucuronidase and distant from the clades of mammalian HAases, in agreement with previous speculations that heparin-like polymers were the first ancestral glucosaminoglycans in metazoan life and that leech HAase possibly evolved to acquire specificity for HA, losing its ancestral heparanase activity18. Both the MSA and phylogenetic tree analysis suggested that LHyal should be a potential endo-β-glucuronidase (family 79), consistent with a previous classification13.
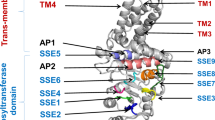
Sequence comparison, structural homology modelling and phylogram of LHyal.
(a) Multiple sequence alignment of LHyal. The figure was generated using the CLUSTALW 1.8 software package based on the known heparanases from Homo sapiens (BAG52128), Ceratotherium simum simum (XP_004430958), Rattus norvegicus (NP_072127) and Mus musculus (NP_690016). The conserved residues are marked with ‘*’ and boxed in yellow, with the exception of the two predicted key catalytic residues, which are coloured red. Semi-conserved sites are boxed in pale blue. The secondary structure predicted by the I-TASSER server is indicated by green arrows to represent β-strands and red cylinders to represent α-helices. (b) Cartoon diagrams of the LHyal 3D structural model. The overall model was constructed using the I-TASSER server based on homology modelling utilising the recently solved crystal structure of β-glucuronidase (PDB code: 3VNY) from Acidobacterium capsulatum. The main catalytic domain is an overall regular (β/α)8 TIM barrel fold (Aβ1-4 surrounded by αH1-8, shown in a), which is typical of glycoside hydrolases. The secondary structure elements in the model are colour-coded (helices in red, β-sheets in yellow, others in green). The deduced catalytic residues, Glu172 and D170, are shown in stick form in purple and blue, respectively (labelled). (c) Cartoon diagrams of the bee venom HAase crystal structure (PDB code: 1FCV). The main catalytic domain is shown as an overall distorted (β/α)8 TIM barrel fold. The active-site residues Glu113 and Asp111 are coloured in purple and blue, respectively. (d) Phylogenetic analysis of the LHyal sequence. The phylogenetic tree of HAase family proteins was constructed using the neighbour-joining (NJ) method. The data are presented as an NJ tree based on the amino acid sequences of LHyal and representative glycoside hydrolases: eight related mammalian HAases, four microbial HAases and three representative glycoside hydrolases from family 79. The lengths of the nodes represent the substitution rate, which is defined as the percentage of substitution sites per alignment length. Bootstrap values >50% are shown on the branch points. One thousand bootstrap replications were performed using the software MEGA5.053.
The mechanism of action of the third group of endo-β-glucuronidases resembles that of the eukaryotic or vertebrate hyaluronate 4-glycanohydrolases more closely than that of the bacterial hyaluronate lyases18. Accordingly, a 3D model of LHyal was built based on the recently solved crystal structures of β-glucuronidase (PDB: 3VNY) from Acidobacterium capsulatum19 using the widely used server I-TASSER, which depends on multiple-threading alignments and iterative template fragment assembly simulations20. The overall fold model suggests that, similar to the well-characterised bee venom hyaluronidase (bvHyal) and human Hyaluronidase-1(hHyal-1)21,22,23, LHyal consists of two distinct structural domains: a potential catalytic domain and a small β-domain (Fig. 1b). Similar to bvHyal (Fig. 1c) and hHyal-123,24, the potential catalytic domain of LHyal adopts a classic and regular (β/α)8 barrel fold and contains the residues Glu172 and Asp170, which are conserved in hyaluronan catalysis23 and are also common to many hydrolases22. The small β-domain adapts a typical antiparallel structure consisting of six β-strands (1 N-terminal β-strand and 5 successive C-terminal β-strands, Fig. 1b); this structure is also commonly observed in many glycoside hydrolases25.
High-level production of functional recombinant LHyal
To prepare to investigate the properties of the potential hyaluronidase LHyal, P. pastoris GS115, a widely used eukaryotic expression system with unique advantages (such as high-density growth, endotoxin-free culture supernatant, secretory expression for easy purification and glycosylation)26,27,28, was selected for the heterologous expression of LHyal. The α-factor signal peptide from Saccharomyces cerevisiae was fused with the LHyal sequence to enable secretory expression. Flask cultivation demonstrated that LHyal was successfully expressed and secreted into medium with a final HAase activity of 11954 U ml−1 (Fig. 2b). A visual HA plate assay was also performed to verify and confirm this HAase activity. HA hydrolysis by the culture supernatant produced a clear transparent zone (Fig. 2a), strongly demonstrating that leech LHyal is an HAase and can be functionally overexpressed in P. pastoris.
Analysis of the functional activity and protein expression level of recombinant LHyal.
(a) Determination of HAase activity using a typical plate assay. Cylindrical holes were injected with the fermentation of LHyal-expressing (left) or control P. pastoris GS115 (right). The clear circle corresponds to the distance diffused by LHyal, while the control (undigested HA) precipitated in the gel upon the addition of cetylpyridinium chloride. (b) Effects of various length N-terminal His-tags on LHyal expression. The gene copy numbers of all recombinant strains were screened with G418 (3 mg ml−1) on YPD plates. Typical time course of recombinant LHyal (C) and H6LHyal (d) expression by P. pastoris in a 3-L fermenter during the methanol induction phase. LHyal production (red  , U ml−1) and cell growth density (green
, U ml−1) and cell growth density (green  , OD600 value) were measured at regular intervals. SDS-PAGE analysis of LHyal (e) and H6LHyal (f) expression in the 3-L fermenter at regular intervals during the methanol induction phase. The protein production of LHyal and H6LHyal is consistent with the enzymatic activity shown in Fig 3c and 3d, respectively. Black arrows indicate the target protein.
, OD600 value) were measured at regular intervals. SDS-PAGE analysis of LHyal (e) and H6LHyal (f) expression in the 3-L fermenter at regular intervals during the methanol induction phase. The protein production of LHyal and H6LHyal is consistent with the enzymatic activity shown in Fig 3c and 3d, respectively. Black arrows indicate the target protein.
His-tags have been extensively applied for recombinant protein expression (generally as a 6xHis-tag) to simplify and facilitate the purification process. Four His-tags of different lengths were fused to the N-terminus of LHyal to determine whether different His-tags would influence the protein expression level. As expected, fusion of the different His-tags resulted in the distinct accumulation of extracellular HAase (Fig. 2b) in the flask cultures. In particular, fusion with the 6×His tag (H6LHyal) significantly increased HAase activity, with a titre of 63180 ± 460 U ml−1. To further improve the expression level, high-cell-density cultivation (HCDC) was conducted in a 3-L bioreactor to evaluate the production of recombinant LHyal and H6LHyal. As expected, LHyal production increased remarkably to a high value of 8.50 × 104 U ml−1 (Fig. 2c), a 7.1-fold increase compared to the flask cultures. In contrast, production of the recombinant H6LHyal fused to the 6×His tag was dramatically improved to 8.42 × 105 U ml−1 (approximate 420 mg L−1) (Fig. 2d), an extremely large increase. In parallel, the accumulation of the recombinant LHyal and H6LHyal was monitored by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) analysis at regular intervals. Compared with LHyal (Fig. 2e), the protein bands corresponding to recombinant H6LHyal increased in intensity with prolonged cultivation time (Fig. 2f). Thus, fusion with 6 His residues at the N-terminus increased the expression level, resulting in much higher HAase activity.
Enzymatic properties of the recombinant H6LHyal
The extracellular H6LHyal was first enriched using a Ni-NTA affinity column. After washing away the background proteins with increasing concentrations of imidazole and subsequent dialysis, the purified protein, which had a specific activity of 1.20 × 106 U mg−1, was analysed by SDS-PAGE. A clear band with an apparent molecular weight of 58 kDa (Fig. 3a) was observed, consistent with the value of 58,684.0 Da (Supplementary Fig. S6) observed by matrix-assisted laser desorption/ionisation quadrupole time-of-flight mass spectrometry (MALDI Q-TOF MS). The optimum temperature for H6LHyal activity was 45°C and the activity decreased significantly above 55°C (Fig. 3b). The activation energy of H6LHyal was further calculated from an Arrhenius plot, which yielded a value of 23.90 kJ mol−1 (Fig. 3b). This value is much lower than that of most other hydrolases, indicating the high HA degradation efficiency of H6LHyal. H6LHyal was stable for 2 h when incubated at 55°C but lost 100% of its activity after incubation for 30 min at 60°C (Fig. 3c). Interestingly, a thermal activation phenomenon was observed when H6LHyal was incubated at temperatures below 60°C. H6LHyal exhibited an optimal pH of 6.5 and remarkable stability over a broad pH range (3.0 to 8.0) (Fig. 3d). H6LHyal was significantly inhibited by Mn2+, Cu2+ and Fe3+ (Supplementary Table S3). The substrate specificity of H6LHyal was compared with that of BTH. In addition to HA, BTH can also hydrolyse chondroitin sulphate (Fig. 3g), while H6LHyal cannot hydrolyse chitin, heparin or chondroitin sulphate (Fig. 3e–3g), validating a previous hypothesis that the substrate specificity of LHyal differs from that of BTH29. Specifically, no differences in temperature stability, metal ion sensitivity, pH stability, or substrate specificity were observed between recombinant H6LHyal and LHyal (data not shown).
Biochemical properties of LHyal.
(a) Purification of the H6LHyal protein using Ni-NTA agarose. M = Marker proteins, C = fermentation supernatant of the control strain P. pastoris GS115, FS = fermentation supernatant of H6LHyal, PE = purified H6LHyal. (b) Effect of temperature on LHyal activity. The enzymatic activity of LHyal was measured at various temperatures in 50 mM citrate buffer (pH 5.5) using HA (1.6 mg ml−1) as the substrate. The logarithm (Ln) of the specific activity (U mg−1) was plotted against the reciprocal of the absolute temperature (T). The activation energy (Ea, kJ mol−1) was calculated from the value of the slope (S) and the general gas constant (R, 8.3 J mol−1 K) according to the Arrhenius equation: Ea = −S × 2.306 × R. (c) After pre-incubating the purified enzyme (0.45 mg ml−1) at 38°C (red  ), 50°C (purple
), 50°C (purple  ), 55°C (blue
), 55°C (blue  ) or 60°C (green
) or 60°C (green  ) for up to 180 min, the remaining enzymatic activity was determined at 38°C in 50 mM Na2HPO4-citrate buffer (pH 5.5). (d) Effect of pH on LHyal activity and stability. The enzyme activity was determined at various pH values at 38°C. The stability was assayed at 38°C in 50 mM Na2HPO4-citrate buffer (pH 5.5) after pre-incubating the purified enzyme (0.45 mg ml−1) for 30 min at 25°C in buffers of pH values from 3.0–8.0. The relative activity is represented as a percentage of the maximum activity. (
) for up to 180 min, the remaining enzymatic activity was determined at 38°C in 50 mM Na2HPO4-citrate buffer (pH 5.5). (d) Effect of pH on LHyal activity and stability. The enzyme activity was determined at various pH values at 38°C. The stability was assayed at 38°C in 50 mM Na2HPO4-citrate buffer (pH 5.5) after pre-incubating the purified enzyme (0.45 mg ml−1) for 30 min at 25°C in buffers of pH values from 3.0–8.0. The relative activity is represented as a percentage of the maximum activity. ( ) 50 mM Na2HPO4-citrate buffer (pH 3.0–6.0); (
) 50 mM Na2HPO4-citrate buffer (pH 3.0–6.0); ( ) 50 mM Na2HPO4–NaH2PO4 buffer (pH 6.0–8.0). Analysis of the degradation of chitin (e), heparin (f) and chondroitin sulphate (g) by HAases using the DNS method. In the control group, the corresponding substrates (2 mg ml−1) were incubated without enzyme in 50 mM citrate buffer (pH 5.5) at 38°C for 30 min. In the LHyal and BTH groups, substrate was incubated with LHyal (1,000 U) or bovine testicular hyaluronidase (BTH, 1,000 U) under the same treatment conditions.
) 50 mM Na2HPO4–NaH2PO4 buffer (pH 6.0–8.0). Analysis of the degradation of chitin (e), heparin (f) and chondroitin sulphate (g) by HAases using the DNS method. In the control group, the corresponding substrates (2 mg ml−1) were incubated without enzyme in 50 mM citrate buffer (pH 5.5) at 38°C for 30 min. In the LHyal and BTH groups, substrate was incubated with LHyal (1,000 U) or bovine testicular hyaluronidase (BTH, 1,000 U) under the same treatment conditions.
Exploration of the hydrolysis process
The hydrolysis mechanisms of the bovine testicular HAase and bacterial HAase have been extensively studied and reviewed14,29. However, although different leech HAases have been extracted and described16,30, the hydrolytic process has remained unclear. The high-level production of recombinant H6LHyal enabled us to explore the hydrolytic process in detail. Under normal conditions (Methods), the degradation products obtained at different incubation times (1, 2, 4 and 8 h) were analysed by hybrid ion trap time-of-flight liquid chromatography/mass spectrometry (LCMS-IT-TOF) in negative ion mode (Supplementary Table S2). Surprisingly, after 1 h of incubation, no small HA oligosaccharides (less than 3,000 Da) were detected (Fig. 4a, 4e), in contrast to results observed for other groups of HAases. Further investigation revealed that all high-molecular-weight HAs were rapidly degraded to low-molecular-weight fragments consisting of approximately 30 saccharide units with an average molecular weight of 2.60 × 104 Da (Supplementary Fig. S5). At 2 h, two prominent ion peaks (Fig. 4b) corresponding to the decasaccharide (HA10) and the octasaccharide (HA8) were detected and identified (Fig. 4f); no larger ion peaks corresponding to other oligosaccharides were detected. When the incubation was extended to 4 h, two new ion peaks corresponding to the hexasaccharide (HA6) and the tetrasaccharide (HA4) appeared, while the peak corresponding to HA10 disappeared (Fig. 4c, 4g), indicating that HA10 was completely digested to HA6 and HA4. Finally, only two peaks corresponding to HA6 and HA4 were detected (Fig. 4d, 4h) after 8 h of digestion. The final products, HA6 and HA4, were further analysed and confirmed by multistage mass spectrometry (Supplementary Fig. S7). Consistent with a previous report17, the Moran-Elson reaction revealed that the HA oligosaccharides contained glucuronic acid at the reducing end31 (Supplementary Fig. S3). Interestingly, no disaccharide (HA2) was detected throughout the degradation process. For comparison, we also investigated the hydrolysis products of BTH. In agreement with previous reports, HA2 was generated at the beginning of hydrolysis (Supplementary Fig. S4a) and the oligosaccharides HA2, HA4, HA6, HA8, HA10, HA12 and HA14 were all simultaneously detected after complete degradation (Supplementary Fig. S4b), demonstrating that the HA degradation processes catalysed by BTH and H6LHyal are quite different.
LC-MS-IT-TOF profile of LHyal-catalysed HA hydrolysis.
The reaction mixture, which contained 1.6 mg of HA and 640 U of HAase per ml in 50 mM citrate buffer (pH 5.5), was incubated at 38°C for 1, 2, 4 or 8 h. At each time point, samples were withdrawn and the reaction was terminated by immersion in boiling water, followed by LC-MS-IT-TOF analysis. The total ion chromatograms of the 1-h (a), 2-h (b), 4-h (c) and 8-h (d) reactions are shown. The ion chromatograms of HA tetrasaccharide (HA4), hexasaccharide (HA6), octasaccharide (HA8) and decasaccharide (HA10) are shown in blue, pink, red and green, respectively. The mass spectra of the 1-h (e), 2-h (f), 4-h (g) and 8-h (h) reactions are shown. The anion m/z of 775.22 [M-H]− (HA4), 576.66 [M–H]2− (HA6), 766.22 [M–H]2− (HA8) and 955.77 [M–H]2− (HA10) are marked with blue, pink, red and green, respectively. (d) Schematic representation of the putative hydrolytic process catalysed by LHyal. LC-MS-IT-TOF, which couples atmospheric pressure ionisation with Ion-Trap (IT) and Time-of-Flight (TOF) technologies and delivers high mass accuracy (ppm) and high mass resolution (10,000 at 1,000 m/z) independent of the MS mode, was employed to analyse the hydrolysis of HA oligosaccharides during the degradation process. Analysis conditions and parameters: electrospray ion source; negative-ion acquisition mode; interface voltage of −3.50 kV; nebuliser gas flow of 1.5 l min−1; CDL temperature of 200°C; block heater temperature of 200°C; detector voltage of 1.60 kV; drying gas at 100 kPa; ion accumulation time of 10 msec; MS range of m/z 300 to 1,500; and MS2 or MS3 ranges of m/z 50 to precursor ion mass. The CID parameters were energy of 50%, collision gas at 100% and 10 repeats. MS data were processed with LCMS solution ver. 3.4 software (Shimadzu).
Discussion
HAases have been the subject of intense research due to their considerable potential for use in biotechnological processes, particularly the pharmaceutical industry. The discovery of new HAases with novel properties is thus an important research area14. Although different types of the third family of HAases, leech HAases, have been isolated and biochemically characterised32, no sequence data are available and little is known about this potentially interesting class of enzymes18. In this work, we cloned a novel leech HAase designated LHyal. In silico analysis demonstrated that LHyal should be classified in glycosidase family 79 and is highly homologous to heparanases, indicating a common ancestor of these enzymes33. Although LHyal is quite different from the two other groups of HAases, the classic (β/α)8 barrel fold and the two amino acid residues (D170 and E172) that serve as the nucleophile and acid/base catalysts that are present in human Hyals-1 and PH-20 are conserved24,34. Functional overexpression of LHyal confirmed its identity as a novel HAase. In addition, the successful high-level expression of HAase is desirable for its broad application in clinical medicine35,36. Despite extensive study37,38, BTH is the sole commercially available HAase39 because the expression levels (activity) of most recombinant HAases are too low to meet practical demands. Here, by employing N-terminal engineering and fed-batch fermentation, a large amount of HAase was obtained, with a crude titre of 8.42 × 105 U ml−1, a value much higher than that of commercial BTH (1,005 U mg−1, 20–25% pure) extracted from bovine testicle40. Compared with the acidic BTH14, the easily purified H6LHyal exhibits superior enzymatic properties (a near-neutral optimum pH of 6.5 and an optimum temperature of 45°C, with excellent temperature resistance). In addition, H6LHyal has superior substrate specificity (Fig. 3g) and has no transglycosidase activity. Consequently, H6LHyal is more suitable for medical applications.
HAs of small molecular mass, particularly HA oligosaccharides composed of 4–18 sugar residues, have attracted much attention for their potential in medical applications5,41,42,43, such as cancer therapy and nerve cell regeneration. Although efficient isolation and purification approaches have been developed15,44,45,46, the preparation of HA oligosaccharides of specific lengths as hydrolysis products of bovine testicular or microbial HAases remains low-yield and a broad spectrum of products is typically obtained (more than 20 different HA oligosaccharides)18,29. In contrast, the novel HAase H6LHyal that was identified and highly overexpressed in this work yields a narrow range of products (Fig. 4d), with glucuronic acid at the reducing end (Supplementary Fig. S3). This product specificity might be due to the preferential digestion of longer HA chains by H6LHyal (Fig. 4i) through a nonprocessive endolytic mode, similar to human HAase 247. Rapid degradation of the long HA chains (HA10) and slow degradation of the short chains (HA8) resulted in the accumulation of shorter HA oligosaccharides (HA8, HA6 and HA4) (Fig. 4b, 4c). The results confirmed that LHyal is a hyaluronate 3-glycanohydrolase (EC 3.2.1.36) without transglycosylation activity, consistent with previous reports13,16,17. Therefore, by controlling the incubation time, specific HA oligosaccharides could be produced in high yield, greatly facilitating downstream purification and improving productivity. The above results suggest that the high-yield novel H6LHyal has great potential for the preparation of specific HA oligosaccharides at the commercial scale.
In conclusion, we successfully cloned the first leech hyaluronate 3-glycanohydrolase family (EC 3.2.1.26) gene and achieved its high-level heterologous overexpression. Compared with commercial BTH, this novel HAase has better substrate specificity and a vastly different hydrolysis mechanism. By controlling the incubation time, different HA oligosaccharides, particularly HA10, HA8, HA6 and HA4, can be selectively generated with high yields. The large-scale expression of H6LHyal and the enzymatic production of specific HA oligosaccharides are not only advantageous for the cosmetic, healthcare and medical industries but will also enable polysaccharide chemical synthesis and cancer therapy research48.
Methods
Bacterial strains, plasmids and growth conditions
The E. coli strain DH5α (Stratagene, La Jolla, CA, USA) and pMD19 (Takara Biotech, Dalian, China) were used as the host and vector, respectively, to clone the HAase gene; the resulting plasmid was used for sequencing. E. coli were grown in Luria-Bertani (LB) medium at 37°C, supplemented with 100 μg ml−1 ampicillin. Pichia pastoris GS115 and pPIC9K (Invitrogen, Carlsbad, CA, USA) were used as the host and vector, respectively, for heterologous expression of the HAase. YPD medium (g l−1: yeast extract 10, peptone 20 and glucose 20) was used for seed culture. Buffered minimal glycerol yeast medium (BMGY, yeast extract 10 g l−1, peptone 20 g l−1, glycerol 10 ml l−1, 100 mM potassium phosphate, 0.34% yeast nitrogen base without amino acids [YNB], 4 × 10−5% biotin, 1% ammonium sulphate) and buffered methanol minimal yeast medium (BMMY, yeast extract 10 g l−1, peptone 20 g l−1, 100 mM potassium phosphate, 0.34% YNB, 4 × 10−5% biotin, 1% ammonium sulphate, 1% methanol) were used for flask cultivation. For fermentation, basal salts medium (BSM) with the following composition was used (g l−1): glycerol 40, K2SO4 18, MgSO4·7H2O 14.9, KOH 4.13, H3PO4 27 and CaSO4·2H2O 0.93, with 4.4 ml l−1 of PTM1 trace metal solution. PTM1 solution contained the following (g l−1): CuSO4·5H2O 6, KI 0.09, MnSO4·H2O 3, H3BO3 0.02, MoNa2O4·2H2O 0.2, CoCl2 0.5, ZnCl2 20, FeSO4·7H2O 65 and biotin 0.2, with H2SO4 5.0 ml l−1. For glycerol-fed batch cultivation and methanol-fed batch cultivation, 50% (w/v) glycerol and pure methanol containing 12 ml L−1 PTM1 solution were used, respectively.
Cloning of the HAase gene LHyal
All primers used in this study are provided in Supplementary Table S1. Total RNA was extracted from the heads of wild leeches using a tissue total RNA extraction kit (Hangzhou Biosci Co., Ltd, China.). cDNA was synthesised in a 20-μl reaction (M-MlV reverse transcriptase 1 μl, RNase Inhibitor 1 μl, 5× first-strand buffer 4 μl, oligo(dT)18 primers 1 μl, 10 mM each dNTP mix 1 μl, total RNA 12 μl) using the M-MlV first-strand RT kit (Hangzhou Biosci Co., Ltd, China). The 3′ end of the HAase gene (LHyal) was amplified with gene-specific degenerate primers (GSDF/GSDR), which were designed based on candidate hyaluronidase cDNAs (GenBank: FP628211.1 and JZ186329.1) similar to heparanases from a Hirudo medicinalis library in the EST database (http://www.ncbi.nlm.nih.gov/nucest/). PCRs were performed in 50-μl volumes using 5 μl of cDNA as the template, 20 pmol of each primer, 10 μl of PCR buffer, 0.25 mM dNTPs and 1 unit of DNA Polymerase under the following conditions: 94°C for 3 min; 35 cycles of 94°C for 30 s, 50–55°C for 30 s and 72°C for 1 min; and 72°C for 10 min. To obtain the 5′ fragment of LHyal, first-strand cDNA synthesis was performed to generate the template using the SMART RACE cDNA Amplification Kit (CLONTECH) according to the manufacturer's instructions. 5′-RACE PCR was performed as previously described using Universal Primer Mix (UPM) and a gene-specific antisense primer (GSPR) designed to the obtained 3′-end sequence of LHyal.
Analysis of the LHyal sequence
To obtain the maximum 5′ sequence, the 5′-ends of 5 separate 5′-RACE clones were sequenced. A putative 1470-bp open reading frame (ORF) corresponding to a potential protein-coding segment was identified based on the sequences of the 3′- and 5′-ends and primers (HAaseF and HAaseR) were designed to amplify the LHyal sequence. The PCR products were extracted from an agarose gel and cloned into the pMD19-T simple carrier for sequencing. Initial comparisons of the translated protein sequences to the protein database were performed using the BLASTP program (http://www.ncbi.nlm.nih.gov/blast/). To confirm its identity, the inferred amino acid sequence of the cDNA was aligned (CLUSTALW) against representative HAase sequences (acquired by BLAST searches of the genetic databases) from mammals, venomous insects and bacteria and mammalian heparanases. The theoretical structure of LHyal was obtained by homology modelling using the I-TASSER webserver20 and the crystal structure of the glycoside hydrolase family 79 β-Glucuronidase from Acidobacterium capsulatum as a template19. The estimated accuracy quality of the modelled structure was 0.71 ± 0.11 (TM-score). To analyse the evolutionary relationship of this leech HAase, a phylogenetic tree of LHyal sequences and known HAases and glycoside hydrolases from the GenBank database was constructed using MEGA 5.0.
DNA manipulation
To construct LHyal-pPIC9K, the HAase-encoding gene was amplified with the primers LHyalF/LHyalR, which introduced EcoRI/NotI restriction sites at the 5′ and 3′ ends of the LHyal gene, respectively. The purified PCR product was digested with EcoRI/NotI and ligated into EcoRI/NotI-digested pPIC9K containing the α-factor signalling sequence, creating LHyal-pPIC9K. To produce the constructs in which various His tags were fused to the N-terminus of LHyal, the primers H4LHyalF/LHyalR, H6LHyalF/LHyalR, H9LHyalF/LHyalR and H12LHyalF/LHyalR were used to introduce 4, 6, 9 and 12 His residues at the N-terminus of LHyal to create H4LHyal-pPIC9K, H6LHyal-pPIC9K, H9LHyal-pPIC9K and H12LHyal-pPIC9K, respectively. The recombinant plasmids were transformed into chemically competent E. coli DH5α prepared using standard CaCl2 methods. Unless otherwise stated, the recombinant plasmids were linearised with SalI and transformed into P. pastoris GS115 by electroporation as described previously49.
Overexpression and high-cell-density fermentation of LHyal
Positive P. pastoris recombinants carrying LHyal were cultivated in YPD medium at 30°C. A 5-ml overnight culture was transferred to 50 ml of BMGY medium and incubated in 500-ml Erlenmeyer flasks rocking at 200 rpm at 30°C. When the yeast culture OD600 reached 4–6, the cells were collected and transferred to 50 ml of BMMY induction medium containing methanol (1% v/v) for 96 h at 30°C.
High-cell-density fermentation was performed in a 3-L fermenter (LiFlus GM BioTRON, Korea) containing 800 ml of BSM supplemented with PTM1 trace salts solution50 with the following parameters: temperature of 30°C, pH 5.5, agitation at 500 rpm and air flow rate of 2.0 vvm. Two 50-ml seed cultures were added to the fermenter and when glycerol was exhausted, the fed-batch phase was initiated by feeding 50% (w/v) glycerol plus 12 ml l−1 PTM1 at index-fed-batch rates of 13.5, 16.2, 19.2, 22.8, 27.2 and 32.4 ml h−1 L−1 for the first 6 h. For the subsequent 6 h, the culture was fed at a constant fed-batch rate of 30 ml h−1 l−1. The methanol induction phase was initiated at 25°C and pH 5.5 with agitation at 1,000 rpm by feeding methanol containing 12 ml l−1 PTM1 at a concentration of 1.8% (v/v) for 3–5 h after the glycerol was exhausted. The methanol feed rate was controlled with a methanol sensor (FC-2000, SuperInfo. Tech. Co. Ltd., Shanghai, China).
Purification of the LHyal protein
The fermentation supernatant and Ni-NTA agarose (Qiagen, Hilden, Germany) were loaded onto a gravity-flow column and incubated for 4 h at 4°C. They were then washed with a stepwise gradient of imidazole (10–60 mM) in phosphate buffer to remove contaminating proteins. The bound N-terminal His-tagged protein was eluted from the column with phosphate buffer containing 500 mM imidazole, dialysed against a gradient of sodium chloride concentrations (300, 100 and 0 mM) in 50 mM phosphate buffer pH 6.0 at 4°C for 24 h. The dialysed protein was lyophilised in a Christ Alpha I-5 instrument (Christ, Osterode, Germany). The purified enzymes were analysed by SDS-PAGE and stained with Coomassie Brilliant Blue G-250. The concentration and total amount of protein were determined according to the Coomassie Brilliant Blue R250 assay method.
Enzymatic characterisation of H6LHyal
HAase activity was determined using the simple plate assay of Richman et al51. The assay plate was prepared with 1 mg ml−1 HA, 1.5% agarose, 50 mM sodium citrate (pH 5.5) buffer, 0.15 M NaCl and 0.02% (w/v) sodium azide. The fermentation was emptied into the cylindrical holes (3 mm in diameter) on the agarose plates, incubated at 37°C for h and covered with 10% (w/v) cetylpyridinium chloride. The formation of a distinct clear halo around the hole indicated HAase activity. Leech HAase activity was quantified by measuring the amount of reducing sugar liberated from HA with the 3,5-dinitrosalicylic acid (DNS) colorimetric spectrophotometric method52. One unit of enzymatic activity was tentatively defined as equal to the reducing power of glucuronic acid (glucose equivalents in micrograms) liberated per hour from HA at 38°C and pH 5.5. Specific activity was defined as units of enzyme per mg of protein. Unless otherwise indicated, the standard enzymatic reaction contained 1.6 mg ml−1 of HA as the substrate and an appropriate amount of H6LHyal in 50 mM pH 5.5 citrate buffer in a total volume of 1 ml and was incubated at 38°C for 10 min. The reaction was stopped by immersion in boiling water for 5 min and then examined using the DNS method. Controls without enzyme were prepared and analysed in an identical manner. The substrate specificities of H6LHyal and BTH were determined using heparin, chondroitin sulphate and chitin. N-acetyl-hexosamine moieties at the reducing ends of oligosaccharides liberated from HA were determined using the Morgan–Elson reaction.
Preparation and analysis of low-molecular-weight HA oligomers
High-molecular-weight HA was prepared at a concentration of 2 mg ml−1 in 50 mM citrate buffer (pH 5.5). The reaction mixture, which contained 1.6 mg of HA and 640 U of HAase per ml, was incubated under standard conditions for 1, 2, 4 and 8 h. The hydrolysis of HA by commercial BTH and the resulting products were investigated under the same conditions. The assays were heated in boiling water for 5 min to terminate the reaction, filtered through a 0.22-μm filter and analysed with LCMS-IT-TOF. A 10-μl aliquot of the sample solution was introduced into the ESI ion source of the mass spectrometer via the autosampler of an HPLC system equipped with a C18 column (150 × 2.0 mm; Shimadzu, Kyoto, Japan). The mobile phases were water and acetonitrile and a 0–20 min linear gradient of 0–12% acetonitrile under a constant flow rate (0.2 ml min−1) was used. Mass spectrometry was performed in negative ion mode with scanning over the m/z range from 100–1,500 at 10 s/scan. Multi-stage mass spectrometry was performed under the same operating conditions.
References
Liu, L., Liu, Y., Li, J., Du, G. & Chen, J. Microbial production of hyaluronic acid: current state, challenges and perspectives. Microb. Cell. Fact. 10, 99 (2011).
Noble, P. W., McKee, C. M., Cowman, M. & Shin, H. S. Hyaluronan fragments activate an NF-κB/I-κBα autoregulatory loop in murine macrophages. J. Exp. Med. 183, 2373–2378 (1996).
Lesley, J., Hascall, V. C., Tammi, M. & Hyman, R. Hyaluronan binding by cell surface CD44. J. Biol. Chem. 275, 26967–26975 (2000).
Rooney, P., Wang, M., Kumar, P. & Kumar, S. Angiogenic oligosaccharides of hyaluronan enhance the production of collagens by endothelial-cells. J. Cell Sci. 105, 213–218 (1993).
Toole, B. P., Ghatak, S. & Misra, S. Hyaluronan oligosaccharides as a potential anticancer therapeutic. Curr. Pharm. Biotechnol. 9, 249–252 (2008).
Zeng, C., Toole, B. P., Kinney, S. D., Kuo, J. W. & Stamenkovic, I. Inhibition of tumor growth in vivo by hyaluronan oligomers. Int. J. Cancer. 77, 396–401 (1998).
Seeberger, P. H. & Werz, D. B. Synthesis and medical applications of oligosaccharides. Nature 446, 1046–1051 (2007).
Urakawa, H. et al. Therapeutic potential of hyaluronan oligosaccharides for bone metastasis of breast cancer. J. Orthop. Res. 30, 662–672 (2012).
Termeer, C. et al. Oligosaccharides of Hyaluronan activate dendritic cells via toll-like receptor 4. J. Exp. Med. 195, 99–111 (2002).
Stern, R., Kogan, G., Jedrzejas, M. J. & Soltes, L. The many ways to cleave hyaluronan. Biotechnol. Adv. 25, 537–557 (2007).
Boltje, T. J., Buskas, T. & Boons, G. J. Opportunities and challenges in synthetic oligosaccharide and glycoconjugate research. Nat. Chem. 1, 611–622 (2009).
Weijers, C. A., Franssen, M. C. & Visser, G. M. Glycosyltransferase-catalyzed synthesis of bioactive oligosaccharides. Biotechnol. Adv. 26, 436–456 (2008).
Meyer, K. & Rapport, M. M. Hyaluronidases. Adv. Enzymol. Relat. Subj. Biochem. 13, 199–236 (1952).
El-Safory, N. S., Fazary, A. E. & Lee, C. K. Hyaluronidases, a group of glycosidases: Current and future perspectives. Carbohyd. Polym. 81, 165–181 (2010).
Tawada, A. et al. Large-scale preparation, purification and characterization of hyaluronan oligosaccharides from 4-mers to 52-mers. Glycobiology 12, 421–426 (2002).
Linker, A., Hoffman, P. & Meyer, K. The hyaluronidase of the leech: an endoglucuronidase. Nature 180, 810–811 (1957).
Linker, A., Meyer, K. & Hoffman, P. The production of hyaluronate oligosaccharides by leech hyaluronidase and alkali. J. Biol. Chem. 235, 924–927 (1960).
Stern, R. & Jedrzejas, M. J. Hyaluronidases: their genomics, structures and mechanisms of action. Chem. Rev. 106, 818–839 (2006).
Michikawa, M. et al. Structural and biochemical characterization of glycoside hydrolase family 79 β-glucuronidase from Acidobacterium capsulatum. J. Biol. Chem. 287, 14069–14077 (2012).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Li, S., Kelly, S. J., Lamani, E., Ferraroni, M. & Jedrzejas, M. J. Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. EMBO. J. 19, 1228–1240 (2000).
Henrissat, B. et al. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. USA. 92, 7090–7094 (1995).
Marković-Housley, Z. et al. Crystal structure of hyaluronidase, a major allergen of bee venom. Structure 8, 1025–1035 (2000).
Chao, K. L., Muthukumar, L. & Herzberg, O. Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis. Biochemistry 46, 6911–6920 (2007).
Henrissat, B. & Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 (1997).
De Schutter, K. et al. Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 27, 561–566 (2009).
Vogl, T., Hartner, F. S. & Glieder, A. New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr. Opin. Biotechnol. 24, 1094–1101 (2013).
Cereghino, J. L. & Cregg, J. M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS. Microbiol. Rev. 24, 45–66 (2000).
Kakizaki, I., Ibori, N., Kojima, K., Yamaguchi, M. & Endo, M. Mechanism for the hydrolysis of hyaluronan oligosaccharides by bovine testicular hyaluronidase. FEBS. J. 277, 1776–1786 (2010).
Yuki, H. & Fishman, W. H. Purification and characterization of leech hyaluronic acid-endo-β-glucuronidase. J. Biol. Chem. 238, 1877–1879 (1963).
Muckenschnabel, I., Bernhardt, G., Spruss, T. & Buschauer, A. Pharmacokinetics and tissue distribution of bovine testicular hyaluronidase and vinblastine in mice: an attempt to optimize the mode of adjuvant hyaluronidase administration in cancer chemotherapy. Cancer. lett. 131, 71–84 (1998).
Hovingh, P. & Linker, A. Hyaluronidase activity in leeches (Hirudinea). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 124, 319–326 (1999).
DeAngelis, P. L. Evolution of glycosaminoglycans and their glycosyltransferases: Implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 268, 317–326 (2002).
Arming, S., Strobl, B., Wechselberger, C. & Kreil, G. In vitro mutagenesis of PH-20 hyaluronidase from human sperm. Eur. J. Biochem. 247, 810–814 (1997).
Frost, G. L., Csoka, T. & Stern, R. The hyaluronidases: A chemical, biological and clinical overview. Trends. Glycosci. Glyc. 8, 419–434 (1996).
Benitez, A. et al. Targeting hyaluronidase for cancer therapy: antitumor activity of sulfated hyaluronic acid in prostate cancer cells. Cancer Res. 71, 4085–4095 (2011).
Reitinger, S. et al. High-yield recombinant expression of the extremophile enzyme, bee hyaluronidase in Pichia pastoris. Protein. Expr. Purif. 57, 226–233 (2008).
Ferrer, V. P. et al. A novel hyaluronidase from brown spider (Loxosceles intermedia) venom (dietrich's hyaluronidase): From cloning to functional characterization. PLoS. Negl. Trop. Dis. 7 (2013).
Borders, C. L. & Raftery, M. Purification and partial characterization of testicular hyaluronidase. J. Biol. Chem. 243, 3756–3762 (1968).
Oettl, M., Hoechstetter, J., Asen, I., Bernhardt, G. & Buschauer, A. Comparative characterization of bovine testicular hyaluronidase and a hyaluronate lyase from Streptococcus agalactiae in pharmaceutical preparations. Eur. J. Pharm. Sci. 18, 267–277 (2003).
Kogan, G., Šoltés, L., Stern, R. & Gemeiner, P. Hyaluronic acid: a natural biopolymer with a broad range of biomedical and industrial applications. Biotechnol. Lett. 29, 17–25 (2007).
Termeer, C. C. et al. Oligosaccharides of hyaluronan are potent activators of dendritic cells. J. Immunol. 165, 1863–1870 (2000).
Torigoe, K. et al. Hyaluronan tetrasaccharide promotes regeneration of peripheral nerve: In vivo analysis by film model method. Brain. Res. 1385, 87–92 (2011).
Blundell, C. D. & Almond, A. Enzymatic and chemical methods for the generation of pure hyaluronan oligosaccharides with both odd and even numbers of monosaccharide units. Anal. Biochem. 353, 236–247 (2006).
Tranchepain, F. et al. A complete set of hyaluronan fragments obtained from hydrolysis catalyzed by hyaluronidase: Application to studies of hyaluronan mass distribution by simple HPLC devices. Anal. Biochem. 348, 232–242 (2006).
Mahoney, D. J., Aplin, R. T., Calabro, A., Hascall, V. C. & Day, A. J. Novel methods for the preparation and characterization of hyaluronan oligosaccharides of defined length. Glycobiology 11, 1025–1033 (2001).
Lepperdinger, G., Mullegger, J. & Kreil, G. Hyal2--less active, but more versatile? Matrix. Biol. 20, 509–514 (2001).
Lepenies, B., Yin, J. & Seeberger, P. H. Applications of synthetic carbohydrates to chemical biology. Curr. Opin. Chem. Biol. 14, 404–411 (2010).
Wu, S. & Letchworth, G. J. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 36, 152–155 (2004).
Clare, J., Rayment, F., Ballantine, S., Sreekrishna, K. & Romanos, M. High-level expression of tetanus toxin fragment C in Pichia pastoris strains containing multiple tandem integrations of the gene. Nat. Biotechnol. 9, 455–460 (1991).
Richman, P. G. & Baer, H. A convenient plate assay for the quantitation of hyaluronidase in Hymenoptera venoms. Anal. Biochem. 109, 376–381 (1980).
Ghose, T. Measurement of cellulase activities. Pure. Appl. Chem. 59, 257–268 (1987).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Acknowledgements
We thank professor Jian Yin (Jiangnan University) for discussion. Financial support was provided by the Major State Basic Research Development Program of China (973 Program, 2012CB720802, 2014CB745103), the National High Technology Research and Development Program of China (863 Program, 2012AA022005, 2011AA100905, 2012AA021201), Program for Changjiang Scholars and Innovative Research Team in University (no. IRT1135), the National Natural Science Foundation of China (31200020), the National Science Foundation for Post-doctoral Scientists of China (2013M540414), the Jiangsu Planned Projects for Postdoctoral Research Funds (1301010B) and the 111 Project.
Author information
Authors and Affiliations
Contributions
P.J. and Z.K. designed the experiments, P.J. and N.Z. performed the experiments. Z.K. and J.C. conceived the project, analysed the data and wrote the paper. G.C.D. analysed the data and wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Sequence of the leech HAase has been deposited in the NCBI Sequence Read Archive and is available through GenBank (Accession Number: KJ026763).
Electronic supplementary material
Supplementary Information
Revised supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Jin, P., Kang, Z., Zhang, N. et al. High-yield novel leech hyaluronidase to expedite the preparation of specific hyaluronan oligomers. Sci Rep 4, 4471 (2014). https://doi.org/10.1038/srep04471
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04471
This article is cited by
-
Fermentative production of enantiopure (S)-linalool using a metabolically engineered Pantoea ananatis
Microbial Cell Factories (2021)
-
Production of Industrial Enzymes via Pichia pastoris as a Cell Factory in Bioreactor: Current Status and Future Aspects
The Protein Journal (2021)
-
Temperature-controlled molecular weight of hyaluronic acid produced by engineered Bacillus subtilis
Biotechnology Letters (2021)
-
Eliminating the capsule-like layer to promote glucose uptake for hyaluronan production by engineered Corynebacterium glutamicum
Nature Communications (2020)
-
High-level constitutive expression of leech hyaluronidase with combined strategies in recombinant Pichia pastoris
Applied Microbiology and Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.