Burrowed deep under the foothills near Palo Alto, Calif., scientists scurried through an underground laboratory, making final preparations for a series of explosions. Their plan: blow up tiny crystals of proteins that could reveal one of nature’s best-kept secrets—how plant photosynthesis turns light into chemical energy. The potential payoff: a step toward unlimited clean power.
It was December 2009, and a sleep-deprived team of researchers and students at SLAC National Accelerator Laboratory had been working nonstop for days to set up this experiment at the world’s most powerful x-ray laser, the Linac Coherent Light Source (LCLS), which accelerates electrons to nearly the speed of light. One group feverishly adjusted injectors that would shoot crystals of proteins into the x-ray beam. Another locked and loaded the injector with fresh crystals of a protein complex called photosystem I, which is key to photosynthesis.
At the end of the two-mile accelerator tunnel, the crystals began their march into the intense laser light. But before each of them exploded, its snapshot was taken with a newly developed scientific technique. Today that method promises to reshape our understanding of biology on the tiniest scale because we can now assemble a rapid sequence of such images—shot in femtoseconds, or millionths of a billionth of a second—into movies.
On supporting science journalism
If you're enjoying this article, consider supporting our award-winning journalism by subscribing. By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
Physicist Richard Feynman once said, “Everything that living things do can be understood in terms of the jigglings and wigglings of atoms.” But never before have we been able to directly see the wiggling of atoms and molecules within living things at this speed. Our method, called serial femtosecond crystallography (SFX), lets us watch high-speed molecular dances that determine how medicines affect diseased cells and how chemical reactions convert energy to different forms.
Already research teams around the world have used SFX to reveal fine details of how an experimental drug regulates blood pressure—paving the way to better hypertension medications. SFX has also shown the structure of the enzyme that destroys red blood cells in sleeping sickness, a fatal disease caused by parasites. And it has yielded the first look at the initial steps during photosynthesis that split water into hydrogen and oxygen.
Back in that underground lab in 2009, the stakes were high as x-ray pulses began to annihilate our carefully formed crystals. Many scientists had said SFX would never work and rejected our requests for funding. But then beautiful images of scattered x-rays began to emerge on computer screens. We still remember our cheers erupting around the room as we watched what would become proof that a new field of x-ray science had been born.
X-Ray vision
Before SFX, scientists made amazing advances in detecting the changes of certain chemical structures, but they could not actually observe the most delicate and complex biological structures in action. In the 1980s, for instance, the late chemist Ahmed H. Zewail invented a way to watch atoms move during chemical reactions using ultrafast pulses of visible laser light. Yet the light’s wavelength was too long to distinguish the tiniest details of protein structure. More recently, dramatic advances in microscope technology have produced near-atomic-resolution images of proteins and viruses. But they are not quick enough to capture rapid reactions such as photosynthesis.
Credit: Lucy Reading-Ikkanda
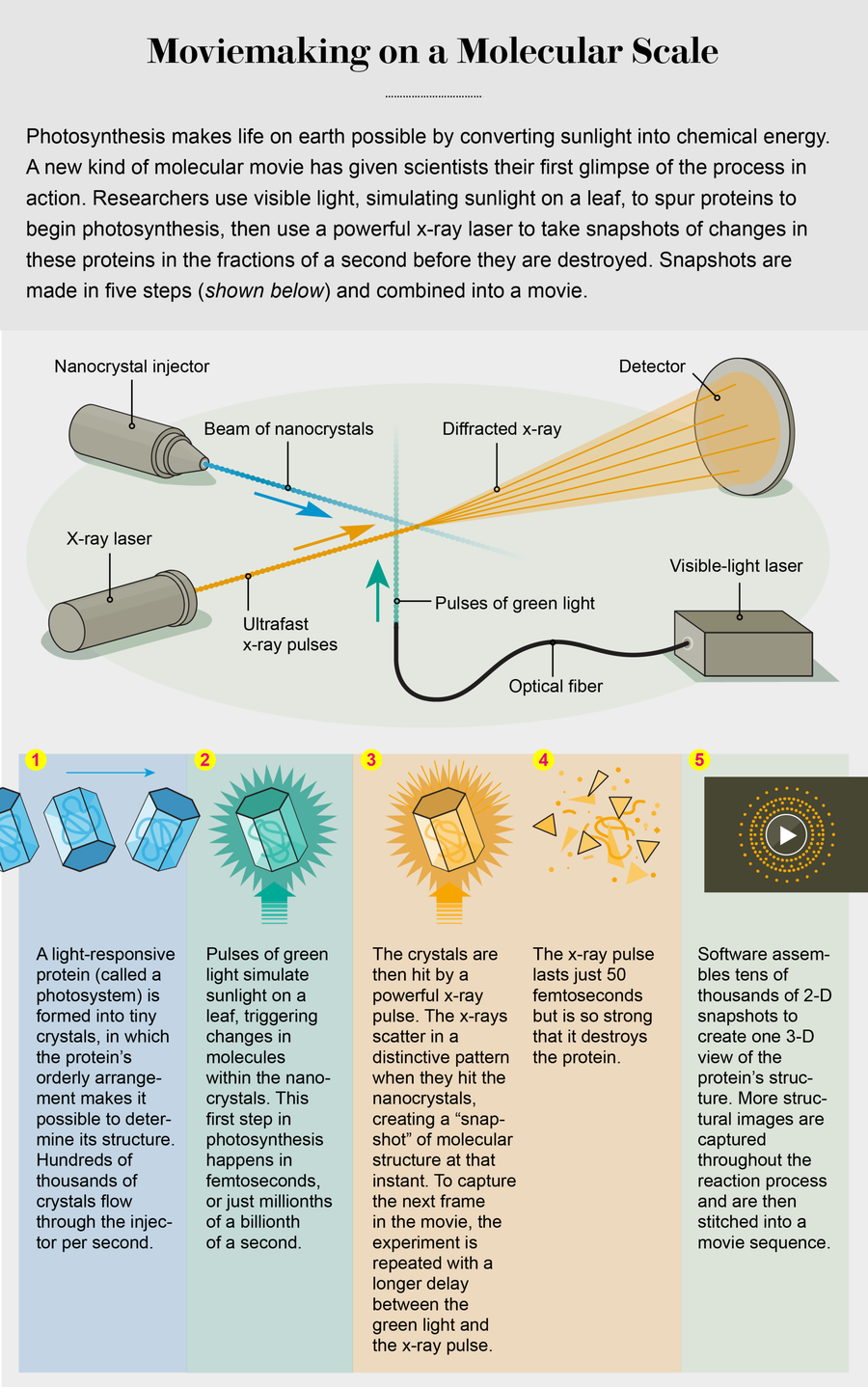
We decided to use x-rays, which have the necessary speed and resolution to record biological reactions in action. Key to our work was developing a technology that would allow x-rays to form pictures of molecules in the instant before destroying them. Traditionally scientists who do this work painstakingly grow large crystals of proteins and other molecules to map the positions of atoms within them. Then they bounce x-rays off the crystals and record the pattern of x-ray scattering, or diffraction. In a crystal, molecules are held in place in an orderly arrangement, so the x-rays scatter in predictable ways, allowing scientists to interpret the position and identity of atoms. This method is called x-ray crystallography, and our serial femtosecond crystallography uses the same principle to see atomic structure but does so far faster.
X-rays ultimately destroy the molecules we are trying to see, however. It was commonly believed that the x-ray laser, which concentrates high-energy x-rays into a powerful beam, would only make matters worse. The laser’s bright light alone can punch a hole through steel. A fragile biomolecule, one would think, would not stand a chance. We needed to outrun the x-rays’ damage and capture an image in femtoseconds. For perspective, the difference between one femtosecond and one full second is equivalent to the difference between a second and 32 million years.
The key to the SFX technique lies in that imperceptible sliver of time between the molecule being struck by the x-ray laser pulse and electrons being ripped off its atoms by x-ray energy. Stripped of electrons, the positively charged remnants repel one another, causing the molecules to expand and ultimately explode.
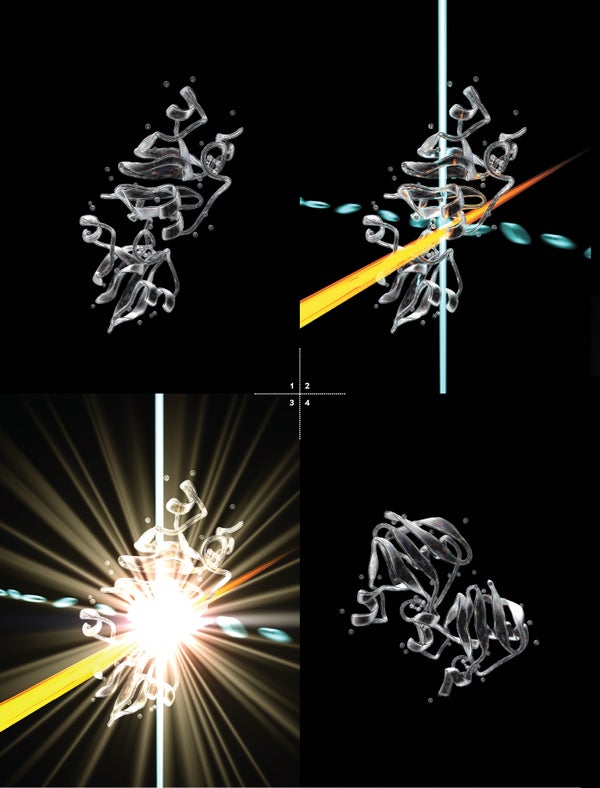
Here is how it works: First, we prompt molecules to interact to form a tiny crystal. Then we shoot a powerful x-ray beam at the crystal in an extremely short pulse, just long enough for some of the x-rays to scatter off the crystal before the beam’s energy rips the molecules apart. Finally, a detector captures the bounced x-rays, whose pattern reveals the type and position of atoms in the protein. By capturing images of a stream of protein crystals as they tumble through the x-ray beam at different angles, we can re-create the structure in 3-D. Finally, we can collect images at different time points in a reaction and put the pictures together in sequence, like images in a film strip.
Crystallized view
The first step toward making these molecular movies came in 2000, when biophysicists Janos Hajdu and Richard Neutze, both then at Uppsala University in Sweden, calculated that it would take roughly 10 femtoseconds for molecules to begin exploding after being hit by x-rays. Thus, scientists needed to take a snapshot faster than that. In 2006 Henry Chapman, now at the German Electron Synchrotron (DESY), and his colleagues were able to do just that using a “diffract then destroy” approach to capture a low-resolution image of two tiny stick figures and the sun etched into a silicon nitride membrane.

SPOT ON: With serial femtosecond crystallography, the gray dots in these panels show patterns of x-rays after they collide with protein crystals, revealing their structure. Credit: Bryan Christie; Source: From “Serial Femtosecond Crystallography Datasets from G Protein-Coupled Receptors,” by Thomas A. White et al., in Scientific Data, Vol. 3, Article No. 160057. Published online August 1, 2016
But would this work for delicate biological molecules? Much of the scientific community was skeptical when we proposed to try. Our first 10 grant proposals were all rejected. Doubters said that the x-ray laser pulses would not be short enough, or the protein crystals would be too small to give any detectable signal, or we would never be able to figure out the crystal’s orientation when it was struck by the x-ray pulse, information needed to determine its structure.
But we thought that if other kinds of molecules could be imaged, as Chapman had proved, then biomolecules could be, too. One of us (Fromme) and her team sought to prove SFX using one of the most difficult tests imaginable: photosystem I. Consisting of 36 proteins and more than 300 light-capturing green and orange pigments, it is among the most complex protein structures analyzed with x-rays to date.
Fromme knew photosystem I intimately, having worked for years to crystallize it and determine its structure using other methods. We also thought that the biomolecular complex’s large size could actually be an advantage because with even a small number of diffraction patterns, we could get a low-resolution image that would be recognizable as photosystem I. And this is what we were able to do in that underground lab in 2009.
Small is beautiful
To get our snapshot, we first needed crystals of photosystem I. In typical crystallography, scientists grow large crystals, which have long been necessary to create enough x-ray scattering to reconstruct a protein’s structure. But it can take years of experimentation to grow large, well-ordered crystals of some proteins. Several have proved nearly impossible, and photosystem I was one of them.
Instead SFX uses nanometer-sized crystals, which are much easier to grow in the lab. Using nanocrystals meant new challenges, however. Not only would we have to get a strong enough signal from such a small crystal, but we faced some basic physical challenges: How do you detect nanocrystals too small to see under a microscope, much less position them in front of x-ray pulses and do so consistently 120 times each second?
First, we had to invent new ways to see our nanocrystals. One of the methods we applied is called SONICC (second-order nonlinear imaging of chiral crystals), in which the crystals convert two ultrafast pulses of infrared light into one green photon—this lights up the nanocrystals like fireflies in the night so that we can detect them.
Another invention shoots the crystals into the x-ray laser pulse at a consistent clip. One of us (Spence), along with physicists Uwe Weierstall of Arizona State University and R. Bruce Doak, now at the Max Planck Institute of Medicine in Heidelberg, Germany, came up with a device that functions much like an inkjet printer, firing a stream of nanocrystal-containing solution across the x-ray beam. This injector fires nanocrystals so precisely that they march into the beam in a single-file line.
To keep the injector from clogging—which could shut down the stream of nanocrystals—Weierstall had to design a wide nozzle that still had the ability to produce a narrow stream. He did this by surrounding the outer end of the nozzle with a stream of helium gas, focusing the stream of crystals to a tiny fraction of a human hair even though the nozzle itself was more than 10 times larger.
Once we had all the machinery in order, we faced one more problem: how to master a Mount Everest of data. A single experiment can generate up to 100 terabytes of data, enough to fill 25 top-of-the-line desktop computer hard drives. And to construct a 3-D view, we have to find, then merge, the correct orientation of each of the crystals in tens of thousands of snapshots. So we developed special software in collaboration with Richard Kirian and Thomas White, both then members of Chapman’s team at DESY. With the new software, we can turn our tsunami of data into accurate 3-D images of a molecule.
Step by step, we improved our technique. And by 2014 our work gave us the first real-time glimpse of the transfer of electrons between two key players in photosynthesis: the large sunlight-catcher photosystem I and a protein called ferredoxin.
When light hits photosystem I, it is converted into electrons, which ferredoxin then carries away to be used for converting CO2 into biological molecules. When ferredoxin leaves, the protein crystals quickly dissolve, making the reaction difficult to follow. Only the superfast process of SFX can see the rapid change.
The next challenge in this line of research is a big focus of Fromme’s work as a biochemist: unraveling how a plant splits water into hydrogen and oxygen using just sunlight and the earth’s abundant metals. Splitting water the way that a plant does could provide cheap, clean-burning hydrogen as fuel for cars and power generators, a long-held dream for developing a renewable energy economy.
We have gathered the first low-resolution snapshots of the water-splitting process and have seen an initial hint of significant structural changes to the protein complex involved: photosystem II. Just recently Jian-Ren Shen’s group working at Okayama University in Japan has applied the SFX technique to show the same snapshot of the process at even more detail. Next, we seek to make high-resolution movies that can reveal all stages of the process at the atomic level and to discover the secret of photosynthesis.
Designer drugs
Now that scientists have begun making movies using SFX, the films we produce could lead not only to future breakthroughs but also more immediately to new and better medications. We saw this potential when we studied angiotensin II receptor blockers (ARBs). These drugs interfere with a cell receptor for the hormone angiotensin II, which constricts blood vessels. ARBs are used to treat high blood pressure (hypertension), the leading cause of stroke and heart failure in the U.S. Whereas the first generation of these drugs has proved useful, the drugs bind to their targets only weakly and must be used in high doses, worsening their side effects, which can include headaches and dizziness and occasionally more serious problems such as swelling in the face and throat.
Our research has revealed the reason behind the poor binding: the drugs really do not fit the receptor as well as they should, so many of their molecules fall away. More accurate structures of the receptors could lead to new ARBs that will more effectively control blood pressure. And in fact, one drug called ZD7155 is already being evaluated.
These refinements could improve many other drugs, too. Angiotensin II receptors belong to a larger and extremely important group of cell receptors called G-protein-coupled receptors. These cell-surface molecules allow a cell to sense and respond to its environment. The scientists who first uncovered the structure and actions of this receptor class won the 2012 Nobel Prize in Chemistry for the breakthrough. The vital role that G-protein-coupled receptors play in cell survival and growth makes them crucial targets for new drugs. Being able to see how their structures change will help pharmaceutical chemists design drugs that fit the receptors precisely and in their active state, thereby reducing the chance of side effects.
“We have shown that in all the previous molecular models, the best guesses for how receptor and drug fit together were wrong in many important details,” says Vadim Cherezov of the University of Southern California, who conducted the angiotensin II experiment. For example, SFX has revealed differences in the structures of G-protein-coupled receptors at room temperature compared with the cryogenically cold temperatures traditionally used in crystallography—meaning that drugs designed for receptors at frozen temperatures will not fit properly when used in the warm human body. (Sometimes drugs hit too broad a target. This is the problem for drugs used to treat sleeping sickness. Our motion pictures have shown that the drugs interact in similar ways with proteins from the parasite that causes the disease and with proteins from human cells. Our more precise images give chemists a chance to make drugs that affect only the parasite protein, not people.)
Eyes have it
We have also been thrilled to see how other researchers are using our SFX techniques to answer different questions. Marius Schmidt of the University of Wisconsin–Milwaukee and his colleagues, for example, have used molecular movies to help explain how our eyes can see. Although we do not typically think of bacteria as being capable of sight, they have light-responsive proteins that are the evolutionary precursors of those in our own visual system. By capturing snapshots faster than ever achieved before, the team made an ultraslow-motion video of extremely rapid events, revealing how a protein in bacteria senses and responds to light.
The group used SFX to capture images of the crystallized protein as it reacted to light in increments of less than a trillionth of a second. Specifically, the team mapped the protein’s atoms in motion as a dye molecule buried within the protein turned yellow in response to light. For the first time, the structure of the yellow dye was captured immediately after it absorbed the light and before it could react; this state is fundamental to light perception in all living organisms, including bacteria and plants, and is the first event in human vision.
Seeing how this protein responds to light not only helps us understand how vision arose but also gives us an unprecedented look at how a biological reaction unfolds on chemistry’s ultrafast timescale. “This puts us dramatically closer to understanding the chemistry necessary for all of life,” Schmidt says.
We are convinced that the future of protein crystallography—as well as our knowledge of nature—lies with the SFX method. And who knows—perhaps within 10 years, half of all known protein structures will not be static images on a textbook page but 3-D movies.