Abstract
The EGR family comprises of EGR 1, EGR 2, EGR 3 and EGR 4 which are involved in the transactivation of several genes. A broad range of extracellular stimuli by growth factors is capable of activating EGR mediated transactivation of genes involved in angiogenesis and cell proliferation. However, their role in controlling VEGF A and FGF 2 signaling in the CL of water buffalo is not known. The present study was conducted to understand the role of EGR mediated regulation of VEGF A and FGF 2 signaling in buffalo luteal cells. Towards this goal, luteal cells were cultured and treated with VEGF A and FGF 2 and the mRNA expression pattern of EGR family members were documented. The EGR 1 message was found to be up-regulated in luteal cells of buffalo at 72 hours of culture. The functional validation of EGR 1 gene was accomplished by knocking out (KO) of EGR 1 in cultured luteal cells by CRISPR/Cas9 mediated gene editing technology. The EGR 1 KO cells were then cultured and stimulated with VEGF A and FGF 2. It was observed that VEGF A and FGF 2 induced angiogenesis, cell proliferation and steroidogenesis in wild type luteal cells, whereas the response of the growth factors was attenuated in the EGR 1 KO cells. Taken together our study provides evidence convincingly that both VEGF and FGF mediate their biological action through a common intermediate, EGR 1, to regulate corpus luteum function of buffalo.
Similar content being viewed by others
Introduction
The corpus luteum (CL) is essential for the pregnancy establishment and its maintenance as it primarily secretes progesterone1. Luteal dysfunction in domestic animals results in inadequate progesterone production, poor embryonic development, infertility and increased pregnancy failure in buffalo cow2. Although, LH and PGF2α plays important role in development and regression of corpus luteum, but there are several other ovarian peptides which are critical for development, function and regression of CL. These include fibroblast growth factors (FGF)3, the vascular endothelial growth factor (VEGF)4, thrombospondins (TSPs)5, IGF6, Leptin7, Angiopoietin8 and Ghrelin9. Among these VEGF helps in the regulation of angiogenesis, vasculogenesis, steroidogenesis of the CL in several species such as cattle10, pigs11, and buffaloes12. The transition of follicular cell to luteal cell, angiogenesis in CL and its function is also controlled by FGF 2 in cattle13 and buffalo3. Growth factors such as VEGF and FGF family mediate their physiological role by binding with their transmembrane tyrosine kinase receptor. The complex dimerizes and stimulates the receptor’s intrinsic tyrosine kinase activity. Ligand activation of Receptor tyrosine kinase (RTK) activates downstream pathways such as mitogen-activated protein kinase (MAPK), protein kinase B (PKB)/ phosphatidylinositol-3-kinase (PI3K) and protein kinase C (PKC) in a ligand and cell-context manner14. The downstream signaling further phosphorylates the dual acceptor motif ERK1/2. Activated ERK translocates into the nucleus and promotes binding SRF/Elk-1 which subsequently upregulates EGR 115,16,17.
FGF signaling showed the transcriptional abundance of NR4A1 and EGR 1 in granulosa cells 18,19. FGF 1, FGF 2 and VEGF are potent EGR genes activators 15,20,21,22. Early growth response genes are key transcriptional regulators which are highly associated with cell growth, survival 23,24 and apoptosis25. The EGR family mainly comprises of four transcriptional factors which includes EGR 1, EGR 2, EGR 3 and EGR 4. Among these transcriptional factors EGR 1, 2 and 3 are activators, whereas EGR 4 acts as a repressor26. EGR proteins bind to GC-rich DNA recognition sites and turn on the expression of different genes required for differentiation and mitogenesis in different tissues27. EGR 1 is also recognized as NGFI-A, KROX24, ZIF268, and TIS8 which is an inducible transcription factor that belongs to a family of immediate early response genes 28,29. EGR 1 is inducibly expressed in many different cell types including endothelial cells in mice30; granulosa cell in bovines31 and luteal cells in bovines32. However, depending on cell type, EGR 1 acts as both a transcriptional activator and repressor33. EGR 1 when binds to DNA it modify gene transcription via its both coactivators and corepressors dependent mechanisms. Since, EGR 1 controls the expression of a wide variety of genes it act as a master regulator23. FGF 1, FGF 2 and VEGF are potent EGR genes activators 15,20,21,22,34. The up regulation of EGR 1 by FGF 2 in endothelial cells suggests its role in inducing proangiogenic factors35. Compelling evidences are provided by Santigo36 which suggests that fibroblast growth factor (FGF 2) should be embodied in the list of critical mediators regulating EGR 1 expression. EGR 1 null mice exhibit impaired reproductive function which shows that EGR 1 is important for the normal functioning of ovaries37. EGR 1 plays a central role in regulation of rapid transition of follicular cells to luteal cells in rats38. According to Liu et al.39, EGR 3 is strongly up regulated by VEGF in human endothelial cells. Various stimuli such as hormones and growth factors also up regulates EGR 3 like EGR 140,41,42.
Although extensive efforts have been made to delineate the mechanisms by which VEGF A and FGF 2 induces angiogenesis, steroidogenesis and cell survivability in murine and human endothelial cells, but no information could be traced to date to understand the insight of EGR mediated regulation of VEGF and FGFs signaling in buffalo luteal cells. Hence, we hypothesized that EGR protein(s) could act as mediators of VEGF and FGFs signaling in bubaline luteal cells. Thus, the present study was aimed to explore the functional role of EGR family members on VEGF and FGF signaling in the luteal cells of water buffalo.
Methods
All methods and experimental protocols were carried out in accordance with relevant safety guidelines and regulations.
Experiment 1
To test whether there is any pairing of expression of EGR genes with VEGF and FGF signaling in bubaline luteal cells.
Collection of CL
Ovaries from healthy buffalo cows with normal reproductive tracts were collected in 1X phosphate-buffered saline (PBS) at 37 °C in a vacuum flask from a local abattoir. In the present experiment, only mid luteal stage corpus luteum was used. The selection of mid stage CL was done according to the pre-estabished protocol43.
Tissue collection, luteal cell culture and treatment with VEGFA and FGF2
The luteal cells were cultured using the pre-established protocol44. In brief, mid stage CLs were removed from the ovary with all connective tissue; and CLs were then sliced up using BP blades (Bard-Parker Surgical Blade). The minced luteal tissue was washed three times at 150 g for 5 min at room temperature with the washing medium containing Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium (Hyclone) and antibiotic–antimycotic solution. The minced cells including luteal, endothelial, pericytes and fibroblasts were then digested by incubating the luteal tissue in (DMEM)/F12 medium containing collagenase, DNase I and bovine serum albumin (BSA) for 2 ×45 min in an orbital shaker incubator at 37 °C. The cells were then filtered through a 70 µm filter and after washing were resuspended in culture medium containing DMEM/F12 medium, 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1% antibiotic–antimycotic solution. The cell viability was determined by Trypan blue vital stain. The cells were then plated out at 1.5 ×105 viable cells per well in a 24-well plate with 1 mL culture medium in a humidified CO2 (5%) incubator at 37 °C. The cells were allowed to attach and grow (75–80% confluent) for 72 hrs (Fig. 1). There were four experimental conditions: (i) Wild type luteal cells; ii) Luteal cell treated with VEGF A @ 100 ng/ml3; (iii) Luteal cell treated with FGF 2 @50 ng/ml3. Cells were cultured for 72 hrs in replicates of six for each group. Spent media was stored at −20 °C until required for P4 RIA and cells were harvested for the total RNA isolation and qPCR analysis to determine the expression pattern of EGR 1, EGR 2, EGR 3, EGR 4, 3β HSD, STAR, CYP11A1, vWF, AKT and BAX genes (Table 1). To assess the cell proliferation and apoptosis MTT and Annexin V apopotosis assay was performed.
Experiment 2
To determine the effect of modulating EGR expression on VEGF and FGF target response in bubaline luteal cells
Production of EGR 1 knock out (KO) luteal cells
Based on the results of Experiment no.1, expression of EGR 1 was found to be up regulated by VEGF A and FGF 2 (Fig. 2) and hence it was chosen to knock out by CRISPR/Cas9 gene editing tool. In this approach, components of CRISPR/Cas9 system (single guide RNA and Cas9) were delivered into the bubaline luteal cells via lipofection and the details of the methodology used are as follows.
Preparation of synthetic single guide RNA (SgRNA)
EGR 1 gene specific guide (20 nt) was designed in silico by using available software like CRISPOR (Table 1). For SgRNA synthesis, the T7 promoter sequence was added to SgRNA template and the in vitro transcription (IVT) template was then generated by polymerase chain reaction (PCR) amplification using designed primers. Briefly, SgRNA DNA template was assembled by PCR using the Phusion High-Fidelity PCR Master Mix (Invitrogen, Cat. No. A29377) in a thermocycler followed by generation of SgRNA by in vitro transcription using the Transcript Aid Enzyme Mix. Later the in vitro-transcribed SgRNA was purified using purification columns kit according to the manufacturer’s instruction. Aliquots from an IVT reaction were separated on an agarose gel to assess the quality of the reaction and the concentration of purified SgRNA was measured by Nanodrop spectrophotometer.
Transfection and genomic cleavage detection
Cultured luteal cells with 30–70% confluency were transfected with Cas9-gRNA RNP by using the Lipofectamine CRISPRMAX Transfection Kit (Invitrogen; Cat. No. CMAX00015) as per the manufacturer’s protocol. Ready-to-transfect wild-type Cas9 protein (GeneArt Platinum Cas9 Nuclease; Cat. No. B25640) was procured from Invitrogen. Briefly, Cas9-gRNA RNP was prepared in one tube and CRISPRMAX reagent diluted in Opti-MEM medium (Cat. No. 11058‐021) in another tube was incubated for 5 min at room temperature. The solution from the tubes was intermixed and incubated further for 10 min and was transfected to luteal cells. Validation of EGR 1 KO was performed using a GeneArt Genomic Cleavage Detection Kit (Invitrogen; Cat No. A24372) as per the manufacturer’s instructions. The cleavage efficiency of the EGR 1 knock out was calculated by the following formulae45.
Treatment of EGR 1 KO luteal cells with VEGFA and FGF2
EGR 1 KO luteal cells were then treated with similar dose of VEGFA and FGF2 as in Experiment no.1 and cultured for 72 hrs in replicates of six in DMEM/F12 for each treatment group. There were five experimental conditions: i) EGR 1 KO luteal cells without treatment; ii) Wild type cells without treatment; iii) EGR 1 KO luteal cells with treatments of VEGFA @ 100 ng/ml; iv) EGR 1 KO luteal cells with treatments of FGF2 @ 50 ng/ml. After 72 hrs of culture, cells were collected from culture plates. The spent media was stored at −20 °C until required for P4 immunoassay and cells were harvested for total RNA isolation and qPCR analysis was performed to determine the expression pattern of EGR 1, 3βHSD, STAR, CYP11A1, vWF, AKT and BAX gene (Table 1).
Cell viability assays
The wild type luteal cells and EGR 1 KO luteal cell were seeded on 96-well plates and incubated at 37 °C for 72 hr with media without treatment and with media containing VEGF A and FGF 2 (100, 50 ng mL−1). Subsequently 20 µl of MTT solution (5 mg/ml in PBS) 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltertrazolium bromide (MTT; MP Biomedical) was added to each well and was incubated for 4 hr at 37 °C. Supernatant was then removed and 100 mL dimethyl sulphoxide (DMSO; MP Biomedical) was added and absorbance at 450 nm was detected with a microplate reader (Biorad).
Apoptosis assay
The differentiation between the apoptotic and healthy cells was performed by Annexin V apoptosis detection kit (BD bioscience). Both wild type and EGR 1 knock out luteal cells were cultured at 37 °C for 72 hrs with media without treatment and with media containing VEGF A and FGF 2 (100, 50 ng mL−1). The cells were trypsinized using 0.05% Trypsin/EDTA solution and were harvested and treated with 5 μl of both annexin V and PI according to manufacturer’s instruction. The apoptotic signal was detected by Axio Observer.Z1 (Carl Zeiss Micro Imaging GmbH, Germany) microscope.
Primers
The primers EGR 1, EGR 2, EGR 3, EGR 4, AKT, BAX, StAR, 3βHSD and CYP11A1 were designed using DNAStar (online trial version, DNASTAR Lasergene 6, 2004), Gene Tool (online trial version, 2004) and Oligo Analyser 3.1 (open access tool, 2017) software. Published primers were used for 40 S ribosomal protein S15 (RPS15A)44 and vWF46. Details of the primers used are presented in Table 1.
Quantitative qPCR analysis
Isolation of total RNA from cultured luteal cells was done by QIAzol reagent (QIAGEN) according to the manufacturer’s instructions. The quality of RNA was determined via A260/A280 in Nanodrop spectrophotometer. Integrity of the total RNA was verified by agarose gel electrophoresis. Total RNA (1 µg) from different samples were reverse transcribed to cDNA using a RevertAid First cDNA synthesis kit (ThermoFisher Scientific) according to the manufacturer’s instructions by using oligo dT primers at 42 °C for 60 min. Quantitative real-time PCR was performed using the Maxima SYBR Green qPCR kit (Thermo Scientific). Each sample was run in triplicate in 25 µL reaction mixture, which consisted of 12.5 µL SYBR green mix, 0.5 µL each of 0.3 µM forward and 0.3 µM reverse primers, 1 µL cDNA and 10.5 µL nuclease-free water. The following general qPCR protocol was followed: initial denaturation at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing and extension at 60 °C for 60 s. The efficiency of Real-time PCR was determined by amplification of a standardized dilution series and slopes.
Hormone determination
Progesterone (P4) concentration in the spent media of cultured luteal cell was estimated by P4125I RIA kit (Immunotech) as per the manufacturer’s instructions. The intra and inter-assay coefficients of variation were 6.5% and 7.2% respectively.
Statistical analyses
All experimental data are shown as mean ± SEM. The statistical significance of differences in mRNA expressions of all genes, P4 concentrations in spent culture media and % cell viability was assessed using the software SPSS.22 (online trial version) by one-way analysis of variance followed by Tukey’s honestly significant difference (HSD) test as a multiple comparison test. Differences were considered to be significant at p < 0.05.
Results
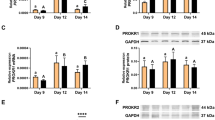
Effect of VEGF A and FGF 2 on expression of Early Growth Response gene in CL
It was observed from the analysis that VEGF A (100 ng/ml) and FGF 2 (50 ng/ml) treatment significantly (p < 0.05; Fig. 2) up regulated EGR 1 gene expression in luteal cells of buffalo at 72 hr. However, the relative expression of other members of EGR family viz. EGR 2, EGR 3 and EGR 4 remained unaffected by the VEGF A and FGF 2 treatment.
Effect of EGR 1 on luteal cell angiogenesis in buffalo
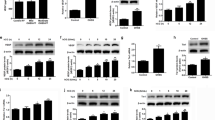
The effect of VEGF A and FGF 2 showed significant up regulation (p < 0.05; Fig. 3A,B) on the expression of the von Willebrand factor (vWF) in treated wild type luteal cells when compared with the wild type luteal cells without treatment. In order to examine the functional role of EGR 1 on luteal cell angiogenesis in water buffalo, EGR 1 was knocked out via CRISPR/Cas 9 genome editing technology (Fig. 4). The present study revealed a significant down regulation in the expression of endothelial cell marker, vWF(p < 0.05; Fig. 3A,B) in EGR 1 KO luteal cells when compared with the wild type luteal cells. Further, to determine the functional role of EGR 1 on VEGF A and FGF 2 induced angiogenesis, the EGR 1 KO luteal cell were treated with VEGF A (100 ng/ml) and FGF 2 (50 ng/ml) over a period of 72 hour. There was a significant (p < 0.05; Fig. 3A,B) difference in the expression of vWF in EGR 1 KO luteal cell with VEGF A and FGF 2 treatment when compared with the wild type luteal cells with treatment. Thus, the present finding confers the angiogenic role of EGR 1 in luteal cells of buffalo.
Gel image of genomic cleavage detection assay of luteal cells transfected with Cas9 and EGR 1 SgRNA using Lipofectamine 2000. Lane 1, 100 bp DNA ladder; Lane 2, negative control sample with intact EGR 1 gene; Lane 3 sample without T7/E1 enzyme digestion, Lane 4: Sample, showing parent and both the cleaved bands after addition of T7/E1 enzyme.
Effect of EGR 1 on luteal cell viability and function in buffalo
To understand the effect of VEGF A and FGF 2 on luteal cell viability, MTT assay was performed. The MTT study revealed significant difference in viability between wild type luteal cells and wild type luteal cell treated with VEGF and FGF 2 (p < 0.05; Fig. 5A,B). The effect of VEGF A and FGF 2 on viability of luteal cells was supported by Annexin V-FITC/PI analysis. After 72 hr incubation, very less green and red fluorescent signals for FITC Annexin and Propidium iodide were detected in wild type luteal cells with treatment compared to that of wild type cells without treatment which shows lesser number of apoptotic and dead cell (Fig. 5C–E) under Axio Observer.Z1 (Carl Zeiss Micro Imaging GmbH, Germany) microscope. Again mRNA expression study of AKT and BAX was done to confirm the effect of VEGF A and FGF 2 in cell proliferation and apoptosis. The study revealed significant up regulation in AKT expression and significant down regulation in BAX (p < 0.05; Fig. 6A,B) expression in wild type treated luteal cells when compared with wild type luteal cells without treatment.
Demonstration of percentage viable n MLC and EGR1 KO MLC treated with VEGF A 100 ng/ml and FGF 2 50 ng/ml at 72 hrs (A,B). Images of wild type luteal cells (C) Wild type luteal cells with VEGF A (D); Wild type luteal cells with FGF 2 (E); EGR 1 KO luteal cells (F); EGR 1 KO luteal cells with VEGF A (G); Wild type luteal cells with FGF 2 (H) showing cells under bright field; Green fluorescence, signal for FITC Annexin; Red fluorescence, signals for Propidium iodide. All the groups of cells were cultured for 72 hour followed by trypsinization and treatment with Annexin V apoptosis detection and Propidium iodide dye.
mRNA expression of AKT and BAX in MLC and EGR1 KO MLC treated with VEGF A 100 ng/ml and FGF 2 50 ng/ml at 72 hrs. All values are shown as mean ± SEM. Different superscripts denote statistically different values (P < 0.05). EGR 1 KO, Early Growth Response factor 1 Knock out; MLC, Mid stage luteal cells.
For understanding the effect of EGR 1 on luteal cell viability and the effect of EGR 1 on VEGF A and FGF 2 induced cell proliferation, MTT assay was performed on EGR 1 KO luteal cells and EGR 1 KO luteal cells with VEGF A and FGF 2 treatment. The study showed significant difference in the viability of EGR 1 KO luteal cells and EGR 1 KO luteal cell with VEGF A and FGF 2 treatment in comparison to the wild type luteal cells with and without treatment. There were almost 70% viable EGR 1 KO luteal cells, 68% viable EGR 1 KO luteal cells with VEGF A treatment and 62% viable EGR 1 KO luteal cell with FGF 2 treatment (p < 0.05; Fig. 5A,B). The effect of EGR 1 on viability was also checked by Annexin V-FITC/PI analysis. After 72 hr incubation, fluorescent signals detected in EGR 1 KO luteal cells and EGR 1 KO luteal cells with VEGF A and FGF 2 treatment were more than wild type luteal cells without treatment and wild type luteal cells with treatment which shows higher number of dead and apoptotic EGR 1 KO cells (Fig. 5F–H). The mRNA expression study of AKT and BAX in EGR 1 KO luteal cells and EGR 1 KO luteal cells with VEGF A and FGF 2 treatment showed significant difference (p < 0.05; Fig. 6A,B) with wild type luteal cells and wild type cells with treatment. These findings indicate that EGR 1 contributes to luteal cell survival and has anti-apoptotic role in VEGF A and FGF 2 signaling in corpus luteum of buffalo.
Effect of EGR 1 on steroidogenesis in luteal cell of buffalo
To determine the steroidogenic role of VEGF A and FGF 2, the progesterone concentration in the spent media of cultured wild type luteal cells and wild type luteal cells with VEGF A and FGF 2 treatment was measured. The treated wild type luteal cells showed significant up regulation (p < 0.05; Fig. 7A,B) in progesterone concentration as compared to the wild type luteal cells without treatment. The relative expression of enzymes controlling the progesterone biosynthetic pathway including StAR, CYP11A1 and 3βHSD was also analysed. The wild type luteal cells with treatment showed significant upregulation of StAR, CYP11A1 and 3βHSD expression in comparison to wild type luteal cells without treatment (Fig. 7C,D). Thus, confirming the steroiodgenic role of VEGF A and FGF 2 in luteal cells of buffalo over a period of 72 hrs.
Progesterone concentration in MLC and EGR1 KO MLC (A,B) and mRNA expression of StAR, CYP11A1 and 3βHSD in MLC and EGR 1 KO MLC treated with VEGF A 100 ng/ml and FGF 2 50 ng/ml at 72 hrs (C,D). All values are shown as mean ± SEM. Different superscripts denote statistically different values (P < 0.05). EGR1 KO, Early Growth Response factor 1 Knock out; MLC, Mid stage luteal cells; 3βHSD, 3beta hydroxyl steroid dehydrogenase; CYP11A1, cytochrome P450 side chain cleavage subfamily A1; StAR, steroidogenic acute regulatory protein.
The effect of EGR 1 on steroidogenesis and its pathway was also studied. The progesterone concentration in the spent media of EGR 1 KO luteal cells did not show any significant difference with the wild type luteal cells (Fig. 7A,B). The expression of steroidogenic enzymes also remains unaffected in the EGR 1 KO luteal cells. However, the EGR 1 KO luteal cells with treatment showed significant difference (p < 0.05; Fig. 7C,D) in progesterone concentration, StAR, CYP11A1 and 3βHSD with the wild type luteal cells with treatment. Thus, the present finding is convincing enough to demonstrate the novel role of EGR 1 in the steroidogenic response to VEGF A and FGF 2 signaling.
Discussion
Early growth response gene stimulates one or more genes which serves as a transcription factor and activates cell-specific response of the genes in order to carry out the ultimate response of the original stimulus. The best known early growth response genes include c-jun, c-fos, and EGR 1. EGR 1 regulates diverse functions including some related to ovarian responses of mitogens, growth factors etc33. It plays a key regulatory role via inducing the expression of genes directing the effector stimuli23. The role of EGR 1 was elucidated in a variety of pathophysiological responses47. In the present study, the relevance of EGR 1 in response to VEGF A and FGF 2 signaling to ovarian physiology has been demonstrated. To the best of our knowledge it is the first study to understand the insight of Early Growth Response genes mediated regulation of FGF 2 and VEGF A signaling in buffalo luteal cells. In the current study, we reported that VEGF A and FGF 2 significantly up regulates EGR 1 mRNA expression (Fig. 2) in wild type luteal cells and causes an EGR 1 dependent transcriptional activation. Previous studies reported the expression of EGR 1 in corpora lutea of cattle 32,48. The mRNA expression of EGR 1 can be enhanced by hypoxia, certain hormones and growth factors like bFGF and VEGF in cultured endothelial cells 49,50. FGF 1, FGF 2 and VEGF are potent EGR genes activators34. EGR 3 is found to have an important downstream role in angiogenesis of VEGF stimulated endothelial cell26. Various stimuli such as hormones and growth factors also up regulates EGR 3 like EGR 140,41,42.
EGR 1 plays a pivotal role in acting as a convergence point for multiple signaling pathways51. VEGF A and FGF 2 stimulates EGR 1 gene by the MAPK signal transduction pathway in many cell types52. According to Santiago et al.36. FGF 2 is an important mediator which regulates the expression of EGR 1 following injury in endothelial cells. FGF signaling increased EGR 1 mRNA levels in bovine granulosa cells18,19. VEGF induces transcriptional abundance of EGR 1 when treated in human microvascular endothelial cells53 and in the uterine stromal cells54.
Angiogenesis is a complex multistep system which includes degradation of endothelial basement membrane, cellular proliferation, canalization, branching and maturation of vessels. Angiogenesis in corpus luteum is regulated by a number of growth factors including IGF6, FGF 23, and VEGF A55. VEGF A and FGF 2 mediate its biological role via binding with trans-membrane tyrosine-kinase receptors, i.e. Flt-1 (VEGFR1) and KDR (VEGFR2) for VEGF 56,57 and FGFR1 to FGFR4 for FGF58. Ligand activation of Receptor tyrosine kinase (RTK) activates MAPK signaling which then phosphorylates ERK1/2 which translocates into nucleus and promotes binding of SRF/Elk-1, which subsequently up regulates EGR 152. EGR 1 is described as the key mediator involved in a wide range of vascular functions59. In order, to validate the transcriptional and functional role of Early Growth Response gene on VEGF and FGF signaling in bubaline luteal cells we have attempted to knockout EGR 1 gene from buffalo luteal cells via a very efficient CRISPR/Cas9 genome editing tool. Previous studies reported that inhibition of EGR 1 inhibits angiogenesis in subcutaneous Matrigel plugs in mice30. We demonstrated significant down regulation of vWF in both the EGR 1 KO luteal cells and EGR 1 KO luteal cell treated with FGF 2 and VEGF A when compared with wild type luteal cells with and without treatment. EGR 1 on activation binds with the GC-rich elements of the promoter regions of genes which are involved in angiogenesis23,30. VEGF induced EGR 1 mediated gene transcription is an important mechanisms for angiogenesis and vasculogenesis processes60. The up regulation of EGR 1 in FGF 2 treated endothelial cells is considered to play an important role in inducing proangiogenic factors35. In another study, EGR 1 has been found additionally engaged with angiogenesis in mouse61.
VEGF A and FGF 2 both play a pivotal role in acting as a cytoprotective factor and inhibiting apoptosis in order to stimulate luteal cell survival 3,4. In the present study, the knocking out of EGR 1 in luteal cells of buffalo showed decrease percentage of cell viability as compared to wild type luteal cells suggesting pro survival role of EGR 1 in luteal cells which can also be confirmed by significant up regulation of BAX and down regulation of AKT in EGR 1 KO cells. When the knock out luteal cell were treated with VEGF A and FGF 2, the percentage of cell viability remained similar to knock out cells without treatment indicating EGR 1 may serve as an important mediator for VEGF A and FGF 2 mediated signaling pathway for cell proliferative responses. Induction of EGR 1 via growth factors in a variety of tissues and cells is immersed in cellular differentiation and proliferation30. EGR 1 protein regulates the expression of genes critical to cell proliferation through direct or indirect mechanism62. EGR 1 has emerged as an important regulator of cellular physiology due to its ability to modulate the expression of cell cycle regulatory proteins 63,64. The embryonic stem cells lacking EGR 1 shows impaired proliferation and differentiation65. EGR 1 is highly associated with cell growth, survival24 and apoptosis25. EGR 1 is a cell proliferation-promoting protein and is also an active part of the apoptotic-signaling cascade, suggesting that it has both pro-survival and pro-apoptotic activities depending on cell context and the primary stimulus47.
VEGF A and FGF 2 play an important role in luteal cell steroidogenesis3,4 which was concluded by the significant up regulation (p < 0.05) of 3βHSD, StAR, and CYP11A1 mRNA expression and P4 estimation in the spent media of wild type luteal cells with VEGF A and FGF 2 treatment. In the present study, our results showed that EGR 1 knock out luteal cells did not show significant difference in the P4 concentration and steroidogenic enzyme expression as compared to the wild type luteal cells. However, the KO luteal cells with treatment showed significant down regulation of expression of key steroidogenic enzyme in comparison to the wild type luteal cells with treatment. In mammals mitogen-activated protein kinase signaling is responsible for the regulation of ovarian steroidogenesis66. The endothelial cells when treated with VEGF A leads to significant up regulation of EGR 1 through activation of MEK/ERK module of MAP67. VEGF and FGF 2 activates downstream signaling pathway involving the mitogen-activated protein kinase (MAPKs) which subsequently converges into EGR 1 promoter35 and the knocking out of EGR 1 led to decreased steroidogenesis. The previous study reported that EGR 1 did not change the progesterone secretion or StAR transcripts expression in bovine granulosa cell31. EGR 1 protein mediates development of follicular and luteal cells. EGR 1 is significant for placental angiogenesis54, embryo implantation and epithelial-stromal cross-talks in the uterus68. In this way, EGR 1 is considered significant for reproductive function and development69. The role of EGR 1 in reproduction was accentuated in genetic studies which has shown that EGR 1 ablated mice were found to be infertile37. Thus, the present study suggests that VEGF A and FGF 2 induced EGR 1 mediated gene transcription is an important mechanisms for steroidogenesis in luteal cells of buffalo.
The results of the present study provide important insights about EGR 1 gene regulation. Our finding confirms that the transcriptional abundance of EGR 1 is induced by VEGF A and FGF 2 and provides a potential link in VEGF and FGF 2 signaling in the cultured luteal cells of buffalo.
Growth factors are considered an essential protein in cell signaling which binds with specific cell surface receptors which further activates and coordinates complex regulatory network functions including cellular proliferation, growth, maturation and their function. Accordingly, they are recognized as beneficial tools in research and biopharmaceutical development with innumerable potential applications. It has been established by many research investigations that two important growth factors viz. VEGF A and FGF 2 regulate ovarian function to a greater extent alone or in combination in water buffalo. Findings of our study suggest that both the growth factors operate through a common intermediate i.e. EGR1 and this paves the way for potential therapeutic application of this identified intermediate in amelioration of reproductive problems in water buffalo.
Conclusion
In the present study, we have demonstrated that VEGF A and FGF 2 significantly up regulates EGR 1 in luteal cells of buffalo and initiates EGR 1 dependent transcriptional activation. The knock out of EGR 1 via CRISPR/Cas9 attenuated VEGF A and FGF 2 induced luteal cell angiogenesis, proliferation and steroidogenesis and also increased apoptosis. The present results characterize the functions of EGR 1 in the regulation of luteal cells angiogenesis, cell proliferation and steroidogenesis and as well as provide important insights into the molecular mechanisms that mediate VEGF A and FGF2 signaling. These results suggest that EGR 1 may function as a part of a multifunctional class of master and control gene regulators.
References
Schams, D. & Berisha, B. Regulation of Corpus Luteum Function in Cattle – an Overview. Reprod Domest Anim 39, 241–251 (2004).
Pate, J. L., Johnson‐Larson, C. J. & Ottobre, J. S. Life or death decisions in the corpus luteum. Reprod Domest Anim 47, 297–303 (2012).
Mishra, S. R. et al. Expression and localization of fibroblast growth factor (FGF) family in corpus luteum during different stages of estrous cycle and synergistic role of FGF2 and vascular endothelial growth factor (VEGF) on steroidogenesis, angiogenesis and survivability of cultured buffalo luteal cells. Agri Gene 1, 53–68 (2016).
Chouhan, V. S. et al. Stimulatory effect of luteinizing hormone, insulin-like growth factor-1, and epidermal growth factor on vascular endothelial growth factor production in cultured bubaline luteal cells. Theriogenology 84, 1185–1196 (2015).
Paul, A. et al. Transcriptional Regulation of Thrombospondins and Its Functional Validation through CRISPR/Cas9 Mediated Gene Editing in Corpus Luteum of Water Buffalo (Bubalus Bubalis). Cell physiol biochem: international journal of experimental cellular physiology, biochemistry, and pharmacology 52(3), 532–52 (2019).
Uniyal, S. et al. Expression and localization of insulin-like growth factor system in corpus luteum during different stages of estrous cycle in water buffaloes (Bubalus bubalis) and the effect of insulin-like growth factor I on production of vascular endothelial growth factor and progesterone in luteal cells cultured in vitro. Theriogenology 83(1), 58–77 (2015).
Kumar, L. et al. Expression of leptin and its receptor in corpus luteum during estrous cycle in buffalo (Bubalus bubalis). Anim Reprod sci 135(1-4), 8–17 (2012).
Mishra, S. R. et al. Expression and localization of angiopoietin family in corpus luteum during different stages of oestrous cycle and modulatory role of angiopoietins on steroidogenesis, angiogenesis and survivability of cultured buffalo luteal cells. Reprod Domest Anim 51, 855–869 (2016).
Gupta, M. et al. Expression and localization of ghrelin and its functional receptor in corpus luteum during different stages of estrous cycle and the modulatory role of ghrelin on progesterone production in cultured luteal cells in buffalo. Domest Anim Endocrin 48, 21–32 (2014).
Berisha, B., Schams, D. & Kosmann, M. Amselgruber, & Einspanier, R. Expression and localisation of vascular endothelial growth factor and basic fibroblast growth factor during the final growth of bovine ovarian follicles. J Endocrinol 167, 371–82 (2000).
Kaczmarek, M. M., Schams, D. & Ziecik, A. J. Role of vascular endothelial growth factor in ovarian physiology: an overview. Reprod Biology 5, 111–36 (2005).
Chouhan, V. S. et al. Expression and localization of vascular endothelial growth factor and its receptors in the corpus luteum during oestrous cycle in water buffaloes (Bubalus bubalis). Reprod Domest Anim 48(5), 810–8 (2013).
Woad, K. J. et al. Fibroblast growth factor 2 is a key determinant of vascular sprouting during bovine luteal angiogenesis. Reproduction 143(1), 35–43 (2012).
Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 103(2), 211–25 (2000).
Damon, D. H., Lange, D. L. & Hattler, B. G. In vitro and in vivo vascular actions of basic fibroblast growth factor (bFGF) in normotensive and spontaneously hypertensive rats. Journal Cardiovasc Pharm 30, 278–284 (1997).
Cholfin, J. A. & Rubenstein, J. L. Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. Journal Comp Neurol 509(2), 144–55 (2008).
Lammi, J. & Aarnisalo, P. FGF-8 stimulates the expression of NR4A orphan nuclear receptors in osteoblasts. Mol cell endocrinol 295(1-2), 87–93 (2008).
Jiang, Z. L., Ripamonte, P., Buratini, J., Portela, V. M. & Price, C. A. Fibroblast growth factor‐2 regulation of sprouty and NR4A genes in bovine ovarian granulosa cells. J cell physiol 226(7), 1820–7 (2011).
Jiang, Z., Guerrero-Netro, H. M., Juengel, J. L. & Price, C. A. Divergence of intracellular signaling pathways and early response genes of two closely related fibroblast growth factors, FGF8 and FGF18, in bovine ovarian granulosa cells. Mol cell endocrinol 375(1-2), 97–105 (2013).
Biesiada, E., Razandi, M. & Levin, E. R. EGR -1 Activates Basic Fibroblast Growth Factor Transcription mechanistic implications for astrocyte proliferation. J Biol Chem 271, 18576–18581 (1996).
Delbridge, G. J. & Khachigian, L. M. FGF−1–Induced Platelet-Derived Growth Factor-A Chain Gene Expression in Endothelial Cells Involves Transcriptional Activation by Early Growth Response Factor−1. Circ Res 81, 282–288 (1997).
Wang, D., Mayo, M. W. & Baldwin, A. S. Jr. Basic fibroblast growth factor transcriptional autoregulation requires EGR −1. Oncogene 14, 2291 (1997).
Khachigian, L. M. & Collins, T. Inducible expression of EGR −1–dependent genes: a paradigm of transcriptional activation in vascular endothelium. Circ Res 81, 457–461 (1997).
Hallahan, D. E. et al. C-jun and EGR −1 participate in DNA synthesis and cell survival in response to ionizing radiation exposure. J Biol Chem 270, 30303–30309 (1995).
Lee, C. G. et al. Early growth response gene 1–mediated apoptosis is essential for transforming growth factor β1–induced pulmonary fibrosis. J Exp Med 200, 377–389 (2004).
Liu, D., Evans, I., Britton, G. & Zachary, I. The zinc-finger transcription factor, early growth response 3, mediates VEGF-induced angiogenesis. Oncogene 27(21), 2989 (2008).
Thiel, G., Müller, I. & Rössler, O. G. Expression, signaling and function of EGR transcription factors in pancreatic β-cells and insulin-responsive tissues. Mol cell endocrinol 388, 10–19 (2014).
Milbrandt, J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science 238(4828), 797–800 (1987).
Cao, X. M. et al. Identification and characterization of the EGR -1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol cell biol 10(5), 1931–1939 (1990).
Fahmy, R. G., Dass, C. R., Sun, L. Q., Chesterman, C. N. & Khachigian, L. M. Transcription factor EGR -1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat med 9, 1026–1032 (2003).
Han, P., Guerrero-Netro, H., Estienne, A., Cao, B. & Price, C. A. Regulation and action of early growth response 1 in bovine granulosa cells. Reproduction 154, 547–557 (2017).
Hou, X. et al. Prostaglandin F2α stimulates the expression and secretion of transforming growth factor B1 via induction of the early growth response 1 gene (EGR 1) in the bovine corpus luteum. Mol Endocrinol 22, 403–14 (2008).
Gashler, A. & Sukhatme, V.P. Early growth response protein 1 (EGR -1): prototype of a zinc-finger family of transcription factors. In Progress in nucleic acid research and molecular biology 50, 191-224 Academic Press (1995).
Yan, S. F. et al. EGR −1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat med 6, 1355 (2000).
Silverman, E. S. & Collins, T. Pathways of EGR −1-mediated gene transcription in vascular biology. The American journal of pathology 154, 665 (1999).
Santiago, F. S., Lowe, H. C., Day, F. L., Chesterman, C. N. & Khachigian, L. M. Early growth response factor-1 induction by injury is triggered by release and paracrine activation by fibroblast growth factor-2. Am J of Pathol 154(3), 937–44 (1998).
Topilko, P. et al. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, EGR −1)-targeted mice. Mol endocrinol 12(1), 107–22 (1998).
Russell, D. L., Doyle, K. M., Gonzales-Robayna, I., Pipaon, C. & Richards, J. S. EGR −1 induction in rat granulosa cells by follicle-stimulating hormone and luteinizing hormone: combinatorial regulation by transcription factors cyclic adenosine 3′, 5′-monophosphate regulatory element binding protein, serum response factor, sp1, and early growth response factor-1. Mol endocrinol 17(4), 520–33 (2003).
Liu, D., Jia, H., Holmes, D. I., Stannard, A. & Zachary, I. Vascular endothelial growth factor–regulated gene expression in endothelial cells: KDR-mediated induction of Egr3 and the related nuclear receptors Nur77, Nurr1, and Nor1. Arterioscl Throm Vas Biol 23(11), 2002–7 (2003).
Levkovitz, Y., O’Donovan, K. J. & Baraban, J. M. Blockade of NGF-induced neurite outgrowth by a dominant-negative inhibitor of the egr family of transcription regulatory factors. J Neurosci 21(1), 45–52 (2001).
Mercier, G., Turque, N. & Schumacher, M. Early activation of transcription factor expression in Schwann cells by progesterone. Mol Brain Res 97(2), 137–48 (2001).
Inoue, A., Omoto, Y., Yamaguchi, Y., Kiyama, R. & Hayashi, S. I. Transcription factor EGR3 is involved in the estrogen-signaling pathway in breast cancer cells. J Mol Endocrinol 32(3), 49–61 (2004).
Sarkar, M., Schilffarth, S., Schams, D., Meyer, H. H. D. & Berisha, B. The expression of leptin and its receptor during different physiological stages in the bovine ovary. Mol Reprod Dev 77, 174–181 (2010).
Paul, A. et al. Regulation of steroidogenic function of luteal cells by thrombospondin and insulin in water buffalo (Bubalus bubalis). Reprod Fert Develop 31(4), 751–759 (2018).
Liang, X. et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 208, 44–53 (2015).
Rajesh, G. et al. Expression and functional role of bone morphogenetic proteins (BMPs) in cyclical corpus luteum in buffalo (Bubalus bubalis). Gen comp endocr 240, 198–213 (2017).
Thiel, G. & Cibelli, G. Regulation of life and death by the zinc finger transcription factor EGR −1. J Cell Physiol 193, 287–292 (2002).
Sayasith, K., Brown, K. A., Lussier, J. G., Dore, M. & Sirois, J. Characterization of bovine early growth response factor-1 and its gonadotropin-dependent regulation in ovarian follicles prior to ovulation. J Mol Endocrinol 37(2), 239–50 (2006).
Ko, Y. et al. Reverse transcriptase—polymerase chain reaction (RT—PCR): a sensitive method to examine basic fibroblast growth factor-induced expression of the early growth response gene-1 (egr-1) in human umbilical arterial endothelial cells. Mol Cell Probes 9(4), 215–22 (1995).
Schwachtgen, J. L., Houston, P., Campbell, C., Sukhatme, V. & Braddock, M. Fluid shear stress activation of egr-1 transcription in cultured human endothelial and epithelial cells is mediated via the extracellular signal-related kinase 1/2 mitogen-activated protein kinase pathway. J Clin Invest 101(11), 2540–2549 (1998).
Tamama, K. & Barbeau, D.J. Early growth response genes signaling supports strong paracrine capability of mesenchymal stem cells. Stem Cells Int (2012).
Pagel, J.I. & Deindl, E. Early growth response 1—a transcription factor in the crossfire of signal transduction cascades (2011).
Szabo, I. L. et al. NSAIDs inhibit the activation of EGR −1 gene in microvascular endothelial cells. A key to inhibition of angiogenesis. J Physiol-Paris 95, 379–383 (2001).
Guo, B. et al. Expression, regulation and function of EGR 1 during implantation and decidualization in mice. Cell cycle 13(16), 2626–2640 (2014).
Chouhan, V. S. et al. Stimulatory effect of vascular endothelial growth factor on progesterone production and survivability of cultured bubaline luteal cells. Anim Reprod Sci 148, 251–259 (2014).
Neufeld, G., Cohen, T., Gengrinovitch, S. & Poltorak, Z. Vascular endothelial growth factor (VEGF) and its receptors. The FASEB journal 13(1), 9–22 (1991).
Zachary, I. VEGF signalling: integration and multi-tasking in endothelial cell biology (2003).
Berisha, B., Steffl, M., Amselgruber, W. & Scham, D. Changes in fibroblast growth factor 2 and its receptors in bovine follicles before and after GnRH application and after ovulation. Reproduction 131(2), 319–329 (2006).
Fu, M. et al. EGR −1 target genes in human endothelial cells identified by microarray analysis. Gene 315, 33–41 (2003).
Lucerna, M. et al. NAB2, a corepressor of EGR −1, inhibits vascular endothelial growth factor-mediated gene induction and angiogenic responses of endothelial cells. J Biol Chem 278, 11433–11440 (2003).
McCaffrey, T. A. et al. High-level expression of Egr-1 and Egr-1–inducible genes in mouse and human atherosclerosis. J Clin Invest 105(5), 653–62 (2000).
Krones-Herzig, A. et al. Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer res 65(12), 5133–43 (2005).
Cheyou, E.R., Youreva, V. & Srivastava, A.K. Involvement of the early growth response protein 1 in vascular pathophysiology: an overview (2014).
Khachigian, L. M. Early growth response-1 in cardiovascular pathobiology. Circ res 98(2), 186–91 (2006).
Lee, S. L., Tourtellotte, L. C., Wesselschmidt, L. & Milbrandt, J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem 270(17), 9971–9977 (1995).
Amsterdam, A., Tajima, K., Frajese, V. & Seger, R. Analysis of signal transduction stimulated by gonadotropins in granulosa cells. Mol Cell Endocrinol 202, 77–80 (2003).
Mechtcheriakova, D., et al. FASEB Jounal 15, 230–242 (2001).
Kim, H. et al. Early growth response 1 (EGR 1) is required for proper epithelial-stromal cross-talks to respond to E2 and P4 for successful embryo implantation in the uterus. Fertil Steril 106(3), e338 (2016).
Lee, S. L. et al. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science 273(5279), 1219–21 (1996).
Acknowledgements
We thank Director of Indian Veterinary Research Institute for providing necessary facilities for conducting this research work.
Author information
Authors and Affiliations
Contributions
M.S. participated in study design, contributed reagents, performed experiments, analyzed data, reviewed and edited the manuscript. S.B. reviewed and revised the manuscript. M.P. performed experiments, analyzed data, wrote the manuscript and prepared the figures. V.S. performed experiments and prepared the figures. A.S. performed experiments and analyzed data. G.S. analyzed data and revised the manuscript. J.G. participated in experimental design and analyzed data. K.W. participated in experimental design and interpreted data. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Punetha, M., Chouhan, V.S., Sonwane, A. et al. Early growth response gene mediates in VEGF and FGF signaling as dissected by CRISPR in corpus luteum of water buffalo. Sci Rep 10, 6849 (2020). https://doi.org/10.1038/s41598-020-63804-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-63804-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.