Abstract
The chordate gastrin/cholecystokinin and ecdysozoan sulfakinin (SK)-signaling systems are functionally and structurally homologous. In the present study, we isolated the cDNA sequences encoding the SK receptors in Rhodnius prolixus (Rhopr-SKR-1 and Rhopr-SKR-2). The Rhopr-SKRs have been functionally characterized and their intracellular signaling pathways analysed via a functional receptor assay. Both Rhopr-SKRs are exclusively activated via the two native R. prolixus sulfakinins, Rhopr-SK-1 and Rhopr-SK-2, but not via nonsulfated Rhopr-SK-1. The Rhopr-SKRs are each linked to the intracellular Ca2+ second messenger pathway, and not to the cyclic AMP pathway. Spatial transcript expression analyses revealed that each Rhopr-SKR is predominantly expressed in the central nervous system with lower expression throughout peripheral tissues. The critical importance of the SK-signaling pathway in the blood-feeding behaviour of R. prolixus was demonstrated by knockdown of the transcripts for Rhopr-SKs and Rhopr-SKRs, which results in an increase in the mass of blood meal taken. The parasite causing Chagas disease is transmitted to the host after R. prolixus has taken a blood meal, and characterization of the SKRs provides further understanding of the coordination of feeding and satiation, and ultimately the transmission of the parasite.
Similar content being viewed by others
Introduction
The chordate gastrin/cholecystokinin (G/CCK) and ecdysozoan sulfakinin (SK) – signaling pathways represent evolutionary divergence of a common ancestral system1. The SKs therefore are considered functionally and structurally homologous to the G/CCK neuropeptides; the earliest discovered gastrointestinal hormones2. Insect SKs, like the G/CCK in chordates, can function as feeding satiety factors. That is to say, once animals have consumed sufficient amounts of food, they become satiated and cease feeding and/or responding to food stimuli3,4. The blood-gorging kissing bug, Rhodnius prolixus is a vector of Chagas disease with the parasite transmitted to humans following the blood meal5. The blood meal is also the trigger for growth, development, and reproduction in R. prolixus. Once per instar, R. prolixus gorges on a blood meal; they do not take another blood meal until the next instar6. In light of the importance of blood-gorging in R. prolixus we sought to examine the SK-signaling pathway and test for its possible influence on the size of blood meal consumed.
The sulfakinin prepropeptide typically codes for two SKs in insects7,8,9,10,11,12,13,14,15,16,17. They are myotropic neuropeptides in a variety of insects, and influence contractions of the hindgut, foregut, and heart2,3,18,19,20,21,22,23,24. Sulfakinins have also been linked to satiety; injection of SK reduced food uptake in R. prolixus17, the red flour beetle Tribolium castaneum25, the blow fly Phormia regina26, the German cockroach Blatella germanica20, and the desert locust Schistocerca gregaria27. Suppression of the SK transcript via RNA interference (RNAi) leads to the stimulation of food intake in Gryllus bimaculatus28 and in T. castaneum29. Knockdown of the two SK receptor transcripts via RNAi in T. castaneum also stimulated food intake25,29.
With regard to G protein-coupled receptors (GPCRs), there are 2 CCK receptors termed CCK receptors 1 (CCK1R) and 2 (CCK2R). Similarly, insects appear to also possess 2 GPCRs for SKs18,29,30. Cholecystokinins may be either sulfated or nonsulfated, leading to variation in their binding affinities to CCK1R and CCK2R. Sulfated CCKs activate CCK1R 500 to 1,000-fold more than the nonsulfated counterpart, whereas both sulfated and nonsulfated CCKs bind to CCK2R in an equivalent fashion30. In insects, although the majority of SKs possess a sulfated tyrosyl residue in their characteristic C-terminal heptapeptide core sequence D/EYGHMRFamide, nonsulfated SKs have been shown to occur in vivo4,9,31. However, the insect SKRs only appear to bind sulfated SKs and not the non-sulfated SKs4,16,21,29.
Despite many studies on SKs in insects, little emphasis has been placed on the SKRs. Insect SKRs have only been functionally characterized in three species, namely D. melanogaster, T. castaneum, and Periplaneta americana. Other insect SKRs have been annotated and, along with the functionally characterized SKRs, belong to the rhodopsin-like family of GPCRs16,29,32,33,34,35,36. Thus far, the intracellular signaling cascades associated with SKRs have only been characterized in D. melanogaster and T. castaneum, and reports related to the intracellular signaling properties of SKRs have been conflicting15,29,36. Zels et al. found that T. castaneum SKRs resemble the signaling properties of the CCK1Rs in vertebrates, which link to both the Ca2+ and cAMP pathways via Gαq and Gαs, respectively. Yu et al. reported that both T. castaneum receptors couple exclusively to the Gαs protein upon activation, stimulating the cAMP pathway29, similar to the D. melanogaster CCKLR-17D135. Another study though found the calcium signaling associated with this receptor to be pertussis toxin (PTX) insensitive, suggesting the D. melanogaster SK receptor CCKLR-17D3 exclusively couples to Gαq/11 proteins16, thereby resembling Type 2 CCKRs30.
In the present study, a functional receptor assay utilizing a human embryonic kidney (HEK)-293 cell line expressing a modified cyclic nucleotide-gated (CNG) channel (HEK293/CNG) was performed to confirm the validity of the two sequenced Rhopr-SK GPCRs, namely Rhopr-SKR-1 and Rhopr-SKR-2, as Rhopr-SK receptors, and to examine the second messenger pathways. Sulfated and nonsulfated Rhopr-SK-1 were tested for the importance of sulfation of the tyrosine residue on ligand binding. Reverse transcriptase quantitative PCR (RT-qPCR) revealed the distribution of the transcripts for Rhopr-SKRs in various tissues in 5th instar R. prolixus, thereby identifying target tissues for Rhopr-SKs. Finally, knockdown of the transcripts for Rhopr-SKs and Rhopr-SKRs were also performed to examine the in vivo role of SKs on the size of blood meal consumed.
Results
Cloning and characterization of Rhopr-SKRs
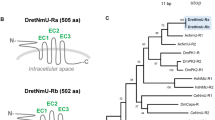
We successfully isolated cDNA sequences spanning 1603 and 1854 base pairs that encode for Rhopr-SKR-1 and Rhopr-SKR-2, respectively. For Rhopr-SKR-1, the open reading frame (ORF) is 1002 base pairs long, encoding a receptor that is composed of 334 amino acids. This ORF is composed of four exons, which are separated by three introns. A poly(A) tail is present in the 3′UTR. Two stop codons are present in the 5′UTR upstream of the start codon, suggesting that the receptor’s ORF is complete. Phosphorylation is predicted at ten amino acid residues. Rhopr-SKR-1 is predicted to contain an extracellular N-terminus, an intracellular C-terminus, as well as seven transmembrane domains. N-linked glycosylation is predicted at one amino acid residue in the extracellular N-terminus (Figs 1A and S1).
Rhodnius prolixus cDNA sequences and the corresponding amino acid sequences of (A) Rhopr-SKR-1 and (B) Rhopr-SKR-2. The numbers of the nucleotides and amino acids (italicized) are to the right of the sequences. For each receptor, the bolded and highlighted methionine is the translation start site. The predicted N-linked glycosylation sites are boxed. The seven predicted hydrophobic transmembrane domains are bolded, highlighted, and underlined. Predicted phosphorylation sites are shaded in grey. Exon boundaries are double-underlined. The ERY and NPITY motifs are italicized and boxed. The 3′ poly(A) tails are underlined.
For Rhopr-SKR-2, the ORF is 1395 base pairs long, which encodes a SK receptor that is composed of 465 amino acids; this ORF is composed of six exons, which are separated by five introns. A poly(A) tail is present in the 3′UTR. Two stop codons are present in the 5′UTR, upstream of the start codon, suggesting that the receptor’s ORF is complete. Phosphorylation is predicted at eight amino acid residues in its intracellular loops. N-linked glycosylation is predicted at two sites at the extracellular N-terminus. This GPCR is predicted to contain an extracellular N-terminus, as well as an intracellular C-terminus, along with seven transmembrane domains (Figs 1B and S2).
Rhopr-SKR-1 and Rhopr-SKR-1 amino acid sequences were aligned via Clustal Omega with a number of their sequenced orthologs from various species, which highlights the high degree of conservation of the transmembrane domains amongst SKRs (Fig. S3). The regions encompassing the seven transmembrane domains of Rhopr-SKR-1 and Rhopr-SKR-2 are highly conserved amongst all of the aligned receptor sequences. The C-termini are also highly conserved, whilst the N-termini contain little conservation. Three of the predicted phosphorylation sites are conserved amongst the receptors, suggesting their functionality. The N-linked glycosylation sites are not conserved amongst the examined receptors (Fig. S3). Rhopr-SKR-1 and Rhopr-SKR-2 display a high degree of similarity, with approximately 43% amino acid identity. Conserved sites that are commonly found in the rhodopsin-like family of GPCRs, such as the intracellular ERY motif (following transmembrane domain III) as well as the NPITY motif (in transmembrane domain VII), are present in both Rhopr-SKR-1 and Rhopr-SKR-2 (Fig. 1).
Functional receptor assays of Rhopr-SKRs
HEK293/CNG cells were utilized to characterize the Rhopr-SKRs and test whether they naturally function via the intracellular Ca2+ (through phospholipase C (PLC)) and/or the cAMP second messenger pathway(s) (see the next section for the second messenger pathways). The PLC pathway in HEK293/CNG cells increases intracellular calcium, which is monitored via luminescence. The CNG channel acts as a biosensor for cAMP, such that an increase in intracellular cAMP levels results in the opening of the CNG channel in the membrane, producing an influx of extracellular calcium into the cell, which is also monitored by luminescence.
The bioluminescence receptor assay confirmed that the two sequenced Rhopr-SKRs, Rhopr-SKR-1 and Rhopr-SKR-2, are SKRs since they are activated by sulfated Rhopr-SK-1 and sulfated Rhopr-SKR-2 in a dose-dependent manner (Figs 2 and 3). HEK293/CNG cells transfected with empty pIRES2-ZsGreen1 plasmids produce no luminescence in response to the addition of Rhopr-SK-1 or Rhopr-SK-2 (not shown), which demonstrates that the luminescence response is due to the presence and activation of Rhopr-SKRs and not due to endogenous receptors present in the HEK293/CNG cells. For Rhopr-SKR-1 and Rhopr-SKR-2, luminescence responses suggest that Rhopr-SK-2 is a partial, but potent, ligand (Figs 2A and 3A). The luminescence response was maximal at 5 seconds following the addition of the Rhopr-SKs for both Rhopr-SKRs (Figs 2B and 3B). For both Rhopr-SKRs, an extended Rhopr-FMRFamide (GNDNFMRFamide), an R. prolixus tachykinin (TK), Rhopr-TK-2, and Rhopr-Kinin-2 did not induce a luminescent response. This indicates that the Rhopr-SKRs are activated by Rhopr-SKs (Figs 2C and 3C). The inability of nonsulfated Rhopr-SK-1 to produce luminescence highlights the importance of the sulfated tyrosyl residue in the peptide’s efficacy when binding to either Rhopr-SKR-1 or Rhopr-SKR-2 (Figs 2A and 3A). For Rhopr-SKR-1, the EC50s for Rhopr-SK-1 and Rhopr-SK-2 are 5.32 × 10−10 M and 8.69 × 10−9 M, respectively (Fig. 2A). For Rhopr-SKR-2, the EC50s for Rhopr-SK-1 and Rhopr-SK-2 are 1.44 × 10−10 M and 1.01 × 10−10 M, respectively (Fig. 3A).
Functional characterization of Rhopr-SKR-1 transiently expressed in HEK293/CNG cells. (A) Dose-response curve showing the activation of Rhopr-SKR-1 via Rhopr-SK-1, Rhopr-SK-2, and ns (nonsulfated) Rhopr-SK-1 as ligands (for the first 5-second interval). EC50 for Rhopr-SK-1 was 5.32 × 10−10 M, whilst the EC50 for Rhopr-SK-2 was 8.69 × 10−9 M. Nonsulfated Rhopr-SK-1 was unable to elicit a response. (B) Kinetics of response activation at 5-second intervals (a total of 15 seconds) at 10−6 M for Rhopr-SK-1 and Rhopr-SK-2. (C) Kinin-2 (Rhopr-Kinin-2), tachykinin (Rhopr-TK-2), and GNDNFMRFamide showed little to no response. Data points are mean ± standard error of the mean for 3 replicates.
Functional characterization of Rhopr-SKR-2 transiently expressed in HEK293/CNG cells. (A) Dose-response curve showing the activation of Rhopr-SKR-2 via Rhopr-SK-1 and Rhopr-SK-2, and ns (nonsulfated) Rhopr-SK-1 as ligands (for the first 5-second interval). EC50 for Rhopr-SK-1 was equivalent to 1.44 × 10−10 M, whilst the EC50 for Rhopr-SK-2 was 1.01 × 10−10 M. Nonsulfated Rhopr-SK-1 was unable to elicit a response. (B) Kinetics of response activation at 5-second intervals (a total of 15 seconds) at 10−6 M for Rhopr-SK-1 and Rhopr-SK-2. (C) Kinin-2 (Rhopr-Kinin-2), tachykinin (Rhopr-TK-2), and GNDNFMRFamide showed little to no response. Data points are mean ± standard error of the mean for 3 replicates.
For both Rhopr-SKRs, the combination of Rhopr-SK-1 and Rhopr-SK-2 resulted in a lower luminescence response when compared to Rhopr-SK-1 alone, indicating that Rhopr-SK-2 competes with Rhopr-SK-1 for both Rhopr-SKRs (Fig. 4).
Luminescence response of Rhopr-SK-1 (1 × 10−6 M), Rhopr-SK-2 (1 × 10−6 M), as well as a mixture of Rhopr-SK-1 and Rhopr-SK-2 (1 × 10−6 M) for (A) Rhopr-SKR-1 and (B) Rhopr-SKR-2. For Rhopr-SKR-1, the observed luminescence was significantly lower for the Rhopr-SK-1 and Rhopr-SK-2 mixture as opposed to Rhopr-SK-1 alone (one way ANOVA followed by Tukey’s Multiple Comparisons Test, ****P < 0.0001). For Rhopr-SKR-2, luminescence was lower for the combination of Rhopr-SK-1 and Rhopr-SK-2 as opposed to Rhopr-SK-1 alone (one way ANOVA followed by Tukey’s Multiple Comparisons Test, P < 0.05), and as opposed to Rhopr-SK-2 alone (one way ANOVA followed by Tukey’s Multiple Comparisons Test, **P < 0.01). Here, luminescence was lower for Rhopr-SK-2 when compared to Rhopr-SK-1 (one way ANOVA followed by Tukey’s Multiple Comparisons Test, ***P < 0.001). Both receptors were transiently expressed in HEK293/CNG cells. Data points are mean ± standard error of the mean for 3 replicates.
Second messenger pathways of Rhopr-SKRs
Incubation of HEK293/CNG cells expressing either Rhopr-SKR-1 or Rhopr-SKR-2 in 10 μM of the PLC inhibitor U73122 abolished the luminescence response to Rhopr-SK-1 and Rhopr-SK-2 (Fig. 5A,B) indicating that the receptors are linked to the PLC second messenger pathway.
Functional characterizations of Rhopr-SKRs in the presence or absence of the PLC inhibitor, U73122 or in calcium-free media containing EGTA. Namely, (A) Rhopr-SKR-1 with U73122, (B) Rhopr-SKR-2 with U73122, (C) Rhopr-SKR-1 with EGTA, and (D) Rhopr-SKR-2 with EGTA. All receptors were transiently expressed in HEK293/CNG cells, with all media containing calcium (in the absence of EGTA). For both Rhopr-SKR-1 and Rhopr-SKR-2, both Rhopr-SK-1 and Rhopr-SK-2 were unable to elicit a response in the presence of 10 μM of U73122 when compared to Rhopr-SK-1 and Rhopr-SK-2 in the absence of U73122. In the absence of U73122 for Rhopr-SKR-1, EC50 for Rhopr-SK-1 was 3.17 × 10−10 M and 1.33 × 10−8 M for Rhopr-SK-2. For Rhopr-SKR-2, EC50 for Rhopr-SK-1 was 1.03 × 10−10 M and 1.21 × 10−10 M for Rhopr-SK-2. In the absence of EGTA for Rhopr-SKR-1, EC50 for Rhopr-SK-1 was 2.94 × 10−10 M and 6.86 × 10−9 M for Rhopr-SK-2. In the presence of EGTA for Rhopr-SKR-1, EC50 for Rhopr-SK-1 was 4.40 × 10−10 M and 3.02 × 10−8 M for Rhopr-SK-2. In the absence of EGTA for Rhopr-SKR-2, EC50 for Rhopr-SK-1 was 2.14 × 10−10 M and 1.34 × 10−10 M for Rhopr-SK-2. In the presence of EGTA for Rhopr-SKR-2, EC50 for Rhopr-SK-1 was 3.44 × 10−10 M and 3.14 × 10−10 M for Rhopr-SK-2. Data points are mean ± standard error of the mean for 3 replicates.
To test if cAMP might also be a second messenger, HEK293/CNG cells expressing either Rhopr-SKR-1 or Rhopr-SKR-2 were incubated in calcium-free media with the extracellular calcium chelator EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid). In these cells, if cAMP is elevated by the application of the ligand, then the CNG channels would open, allowing extracellular calcium to enter the cell and be monitored by luminescence. Calcium-free media (containing EGTA to chelate all of the extracellular calcium) would therefore abolish this effect of cAMP. Figure 5C,D shows that calcium-free media containing EGTA does not abolish the luminescence induced by Rhopr-SK-1 and Rhopr-SK-2, indicating that neither Rhopr-SKRs are linked to the cAMP second messenger pathway. Thus, both Rhopr-SKR-1 and Rhopr-SKR-2 solely function via the PLC/IP3 pathway, elevating intracellular Ca2+ (and not via cAMP). Addition of forskolin as well as dibutyryl-cAMP elicited luminescence responses in cells expressing Rhopr-SKR-1, thereby confirming the presence and functionality of the CNG channels in the HEK293/CNG cells (Fig. S4). Addition of ATP in all conditions elicited the expected positive bioluminescent response, whilst cells in media alone did not elicit luminescence (not shown).
Spatial expression analyses of Rhopr-SKR-1 and Rhopr-SKR-2 via RT-qPCR
Transcript expression levels for Rhopr-SKR-1 were highest in the CNS, with lower levels of expression in the male and female reproductive tissues, the heart, Malpighian tubules, salivary glands, fat body, and gut tissues (Fig. 6A). Transcript expression levels for Rhopr-SKR-2 were also highest in the CNS, with relatively lower levels of expression in the heart, salivary glands, Malpighian tubules, as well as the male and female reproductive tissues, gut, and fat body tissues (Fig. 6B).
Spatial expressions of the (A) Rhopr-SKR-1 and (B) Rhopr-SKR-2 transcripts in Rhodnius prolixus 5th instars via reverse transcriptase quantitative PCR (RT-qPCR). For each receptor, expression was examined in the CNS, foregut, anterior midgut, posterior midgut, hindgut, salivary glands, heart, Malpighian tubules, fat body, male reproductive system, and female reproductive system. Data points are mean ± standard error of the mean for 3 independent samples.
Knockdown of Rhopr-SKs and Rhopr-SKRs via double stranded RNA (dsRNA)
RT-qPCR demonstrated the efficiency of Rhopr-SK dsRNA, whereby the Rhopr-SK expression levels were reduced in 5th instar CNS at day 2 post-injection by 96% (n = 3), expressed as the percentage of knockdown in relation to CNS transcripts from insects injected with the double stranded ampicillin resistance gene (dsARG). For the Rhopr-SKR-1 and Rhopr-SKR-2 dsRNA experiment, expression levels of the transcripts at day 2 post-injection were reduced by 93.1% and 96.8%, respectively (n = 3).
Immunohistochemistry was utilized to verify the efficiency of Rhopr-SK dsRNA knockdown in vivo. Since the Rhopr-SK transcript codes for both Rhopr-SK-1 and Rhopr-SK-2, the reduction or absence of SK-like immunohistochemical staining in insects pre-injected with Rhopr-SK dsRNA confirms that both Rhopr-SKs have decreased expression in the R. prolixus CNS. SK-like staining was drastically reduced in CNS tissues from animals injected with dsRNA (dsRhopr-SK) in relation to insects injected with dsARG (Fig. 7).
Verification of Rhopr-SK knockdown via immunohistochemical staining of the brain in 5th instar Rhodnius prolixus. Insects were injected with the dsRNA of either (A) ARG (dsARG) or (B) Rhopr-SK (dsRhopr-SK) then dissected and examined two days post-injection. Animals injected with dsARG display bright immunoreactive staining throughout the cells and processes of the brain, whereas Rhopr-SK dsRNA-injected animals display reduced staining throughout the same regions. Scale bars: 100 μm.
Silencing of the Rhopr-SKs led to an increase in the overall intake of the blood meal by 31.4%, compared to control animals injected with dsARG. Moreover, the simultaneous silencing of the Rhopr-SKR-1 and Rhopr-SKR-2 led to an increase in the intake of the blood meal by 31.5%, compared to animals injected with dsARG. Such results demonstrate the role of the Rhopr-SK-signaling pathway in satiety in R. prolixus in vivo.
Over the five hours post-feeding, the rate of diuresis (as judged by the loss of weight) was similar between the dsRhopr-SK and dsARG-injected animals, as well as between dsRhopr-SKR-1 + dsRhopr-SKR-2 and dsARG-injected animals, suggesting that Rhopr-SKs and their receptors do not play any direct roles in the rate of diuresis in R. prolixus (Fig. 8).
Post-feeding weights of 5th instar Rhodnius prolixus previously injected (48 hours earlier) with (A) 1 μg of Rhopr-SK dsRNA (dsRhopr-SK) or 1 μg of ARG dsRNA (dsARG); or with (B) a mixture of 1 μg of Rhopr-SKR-1 and 1 μg of Rhopr-SKR-2 dsRNA (dsRhopr-SKRs) or 2 μg of ARG dsRNA (dsARG). Post-feeding weights were significantly lower in control insects injected with ARG dsRNA in comparison to insects injected with either Rhopr-SK dsRNA (two way ANOVA followed by Sidak’s Multiple Comparisons Test, ****P < 0.0001) or Rhopr-SKR-1 with Rhopr-SKR-2 dsRNA (two way ANOVA followed by Sidak’s Multiple Comparisons Test, ****P < 0.0001). Histograms are mean ± standard error of the mean for 25 insects (for each of A,B).
Discussion
In this study, the full coding sequences for both Rhopr-SKR-1 and Rhopr-SKR-2 were cloned and characterized for the first time in the kissing bug R. prolixus. Previously, insect SKRs have only been sequenced in P. americana34, T. castaneum29, and D. melanogaster15,35. The seven hydrophobic transmembrane domains of both Rhopr-SK GPCRs display high sequence similarity to those of other sequenced and predicted SKRs, all of which contain an extracellular N-termini and an intracellular C-termini16,25,29,34,36,37. Conserved signature sequences that are found in the rhodopsin-like family of GPCRs and that are essential for receptor activation are also present in both Rhopr-SKR-1 and Rhopr-SKR-2, including the intracellular ERY motif downstream of transmembrane domain III, as well as the NPITY motif that is within transmembrane domain VII. The amino acid sequences of the Rhopr-SKRs exhibit a high degree of sequence homology with one another, displaying 43% similarity. In relation to their human homologs CCK1R and CCK2R, Rhopr-SKR-1 displays a higher degree of sequence homology with CCK1R (40.5%) than with CCK2R (35.8%), whilst Rhopr-SKR-2 displays an approximately equal degree of sequence homology with both CCK1R and CCK2R (33.5% and 33.4%, respectively).
The signaling properties of both Rhopr-SKR-1 and Rhopr-SKR-2 were analyzed in this study. Both Rhopr-SKR-1 and Rhopr-SKR-2 were activated by sulfated Rhopr-SK-1 and sulfated Rhopr-SK-2 in a dose-dependent manner, indicating that these two peptides are ligands for the receptors. In addition, the results indicate that both Rhopr-SKR-1 and Rhopr-SKR-2 are linked to the intracellular Ca2+ pathway via Gαq, and do not act via the cAMP pathway. Rhopr-SKRs’ stringent Rhopr-SK agonist requirements for their activation was also demonstrated as members of other families of peptides, Rhopr-Kinin-2, Rhopr-TK-2, and an extended Rhopr-FMRFa, were unable to elicit a response. This study also highlights the importance of the sulfated tyrosyl residue in the efficacy of Rhopr-SKs when binding to either Rhopr-SKR-1 or Rhopr-SKR-2, as nonsulfated Rhopr-SK-1 was unable to activate either of the Rhopr-SKRs. Thus, the sulfated tyrosyl is required for optimal binding and activation of Rhopr-SKRs by Rhopr-SKs.
Although the results of our in vitro characterization of the Rhopr-SKRs and their second messenger pathways might not reflect all in vivo SK-signaling pathways, they are in line with some of the earlier findings regarding SKRs. Mammalian CCKRs and nematode CCK-like receptors have been typically found to function through the heterotrimeric G proteins of the Gαq family, with only CCK1R also coupling to Gαs30,38,39,40,41. Such findings mainly align with our results, whereby both Rhopr-SKRs were found to couple to Gαq-dependent signaling pathways. For Rhopr-SKR-1, Rhopr-SK-1 (EC50 = 5.32 × 10−10 M) was found to be one order of magnitude more potent than Rhopr-SK-2 (EC50 = 8.69 × 10−9 M). For Rhopr-SKR-2, the potency of Rhopr-SK-1 (EC50 = 1.44 × 10−10 M) and Rhopr-SK-2 (EC50 = 1.01 × 10−10 M) were similar. The dose response curves demonstrate that Rhopr-SK-2 serves as a partial, though potent ligand whereas Rhopr-SK-1 acts as a full, and potent ligand in activating the Rhopr-SK receptors. The unique carboxyl-terminal amino acid sequence of Rhopr-SK-2, whereby the histidine is replaced with a tyrosyl residue in the sequence YGHMRFamide, might explain the partial ligand activity in relation to the highly conserved sequence of Rhopr-SK-1.
The transcript profiles of both Rhopr-SKR-1 and Rhopr-SKR-2 demonstrate that expression for both receptors is highest in the CNS, with lower levels of expression in peripheral tissues. Similar distributions of SKRs were found in T. castaneum, where expression levels for both of the transcripts were predominantly in the head region25,29. Zels et al. have found that both SKR-1 and SKR-2 are mainly present in the brain and optic lobes of T. castaneum, with lower expression in the salivary glands, gut, fat body, testes, and ovaries36. In humans, CCK1R is predominantly expressed in the alimentary tract, whereas CCK2R is mainly expressed in the brain42. The widespread expression of both Rhopr-SKRs in peripheral tissues indicates that Rhopr-SKs might serve as neurohormones that circulate in the hemolymph, influencing a variety of tissues. Mammalian CCKRs have been linked to various physiological processes that include, but are not limited to, the regulation of food intake. In humans, CCK1R has been linked to additional processes, which include stimulating pepsinogen secretion and slowing gastrointestinal motility43, whilst CCK2R has been linked to memory, anxiety, and the secretion of gastric acid44. Additionally, insect SKs have also been linked to synaptic growth35, locomotion, odor preference45, and release of digestive enzymes46,47,48,49. In P. americana, RT-qPCR demonstrated the presence of an SKR in the CNS, and immunocytochemistry demonstrated the receptor’s presence in gut membrane fractions34. The transcript expression of Rhopr-SKRs in the reproductive tissues in 5th instar R. prolixus suggests their potential role in reproductive processes such as egg production as well as spermatogenesis. In the insect Zophobas atratus, SK-1 was found to inhibit spontaneous oviduct and ejaculatory duct contractions24,50. The presence of the Rhopr-SKRs in the fat body indicates potential involvement of SKs in energy storage and release, and indeed, Slocinska et al. found that SK-1 affects the concentrations of proteins, lipids, and glycogen in the larval fat bodies of Z. atratus50.
Silencing of SKs via RNAi has been linked to stimulation in the intake of food in G. bimaculatus28 and T. castaneum29. Additionally, RNAi-induced silencing of each of the transcripts for the two SKRs in T. castaneum led an increase of food intake by approximately 20% for SKR125 and 50% for SKR229. Through the use of a feeding assay, we previously demonstrated the role of Rhopr-SKs in the regulation of food intake in R. prolixus17, whereby insects injected with Rhopr-SK-1 took smaller blood meals. In the present study, the transcript coding for Rhopr-SK-1 and Rhopr-SK-2 as well as the transcripts coding for Rhopr-SKR-1 and Rhopr-SKR-2 were successfully silenced via injection of their dsRNA into 5th instar R. prolixus. Silencing expression of the Rhopr-SKs and the Rhopr-SKRs in R. prolixus led to an increase in the overall size of the blood meal consumed by 31.4% and 31.5%, respectively.
In conclusion, the cDNAs of Rhopr-SKR-1 and Rhopr-SKR-2 have been cloned and functionally characterized. Knockdown results provide strong evidence that the Rhopr-SK-signaling pathway is involved in the regulation of food intake in the kissing bug R. prolixus, particularly in satiation. The wide spatial distribution of the two Rhopr-SKRs highlights their importance in modulating feeding-related processes as well as other potential roles in processes such as energy metabolism and reproduction. The two Rhopr-SKRs, which are activated by both sulfated Rhopr-SKs, function exclusively via the intracellular Ca2+ second messenger pathway and not via the cAMP second messenger pathway. These findings regarding the localization and signaling pathways of the Rhopr-SKRs contribute to future studies regarding potential applications of SKs in pest management. R. prolixus is the vector of Chagas disease, with the parasite passed on to humans following blood-feeding. Interfering with the Rhopr-SK-signaling pathway would impact blood-feeding and thereby interfere with the transmission of Chagas disease.
Materials and Methods
Animals
Male and female 5th instar R. prolixus were obtained from a colony raised at the University of Toronto Mississauga. Insects were reared at 50% humidity, 25 °C in incubators and fed defibrinated rabbit blood (Cedarlane Laboratories Inc., Burlington, ON, Canada), once in each instar. All tissues were dissected from unfed 5th instars, 3 to 4 weeks after ecdysis into 5th instars.
Chemicals
Rhopr-SK-1 (pQFNEY(SO3H)GHMRFamide) and Rhopr-SK-2 (GGSDEKFDDY(SO3H)GYMRFamide) were custom synthesized by SynPeptide (Shanghai, China) at >95% purity, and the sequences including the sulfation were confirmed by the Advanced Protein Technology Centre (Hospital for Sick Children, Toronto, ON). Rhopr-FMRFamide (GNDNFMRFamide), Rhopr-tachykinin-2 (Rhopr-TK-2) (APSTMGFQGVRamide), and Rhopr-Kinin-2 (AKFSSWGamide) were custom synthesized by GenScript (Piscataway, NJ, USA) at >95% purity. All peptides were reconstituted in double-distilled water into stock solutions at 10−3 M. In the physiological assays, stock solutions were stored at −20 °C as 10 μl aliquots until working solutions were made using physiological saline (KCl 8.6 mM, NaCl 150 mM, CaCl2 2 mM, NaHCO3 4 mM, glucose 34 mM, MgCl2 8.5 mM, HEPES 5 mM [pH 7.0]). An anti-perisulfakinin antibody was provided by Jan Veenstra (Bordeaux, France), with its specificity described by Veenstra et al.51. Goat cyanine dye 3 (Cy3) anti-rabbit (IgG) secondary antibody was purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA). Both antibodies were stored at −20 °C. Triton X-100, forskolin, dibutyryl cAMP, and U73122 were obtained from Sigma Aldrich, Oakville, ON, Canada.
Cloning and characterization of the transcripts for Rhopr-SKRs
Sulfakinin receptors 1 and 2, Rhopr-SKR-1 and Rhopr-SKR-2, were isolated and characterized from 5th instar R. prolixus. Here, the putative, partial, amino acid sequences for the Rhopr-SKRs, which were previously annotated by Ons et al.52, were used to search the R. prolixus genome and transcriptome assemblies via BLAST53. The remaining in silico procedures were performed in Geneious v. 4.7.6 (http://www.geneious.com). Using the results, putative sequences of the SK receptors were highlighted in the R. prolixus genome. Then, 5′ and 3′ rapid amplification of cDNA ends (RACE), as well as a modified 5′ and 3′ RACE technique, was applied to obtain the full open reading frames (ORFs) of Rhopr-SKR-1 and Rhopr-SKR-2 (Thermo Fisher Scientific, Waltham, MA, USA). A library plasmid DNA (pDNR-LIB plasmid) of the central nervous system (CNS) that was previously prepared by Paluzzi et al. was used as a template for the modified 5′ and 3′ RACE PCRs54. A variety of gene-specific as well as plasmid-specific forward and reverse primers were utilized for the modified 3′ and 5′ RACE reactions. The primers were used to amplify specific products using a nested PCR approach (Table S1). All reactions were performed using an s1000 thermal cycler (BioRad Laboratories, Mississauga, ON, Canada). The PCR thermal cycler conditions were maintained at an initial 3 minutes at 94 °C, which was followed by 30 cycles of 30 seconds at 94 °C, 30 seconds at an annealing temperature appropriate to the utilized primers, and 60 seconds at 72 °C. The final step consisted of running the samples for 10 minutes at 72 °C. Following each PCR reaction, the resulting amplicon/template cDNA were gel purified using the EZ-10 Spin Column DNA Gel Extraction Kit (Bio Basic Inc., Markham, ON) then used as a template for the following (nested) PCR reaction. Subsequently, the final RACE PCR products were gel purified and ligated onto a pGEM T Vector System (Promega Corporation, Madison, WI, USA). The amplified cDNA-containing vectors were extracted from the cells via the EZ-10 Spin Column Plasmid DNA Miniprep Kit (Bio Basic Inc., Markham, ON), and sequenced at The Hospital for Sick Children’s DNA sequencing facility (Toronto, ON, Canada) or at Eurofins Canada (North York, ON, Canada). To ensure the accuracy of the obtained sequences, sequencing results were confirmed via several independent clones (minimum of 3).
Sequence analysis of Rhopr-SKRs
The transmembrane domains for the Rhopr-SKRs were predicted via TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Intron-exon boundaries were found via a BLAST search using the R. prolixus genome assembly and confirmed using the online splice site prediction tool (http://www.fruitfly.org/seq_tools/splice.html). NetPhos 3.1 server was used to predict the potential intracellular phosphorylation sites (http://www.cbs.dtu.dk/services/NetPhos/), and NetNGlyc 1.0 server for predicting the potential N-linked glycosylation sites (http://www.cbs.dtu.dk/services/NetNGlyc/). Alignments of the orthologous receptor sequences were performed via Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Boxshade 3.21 server (http://www.ch.embnet.org/software/BOX_form.html) was used to obtain a multiple sequence alignment figure.
Mammalian expression vectors and transfection of Rhopr-SKRs
The ORFs of Rhopr-SKR-1 and Rhopr-SKR-2 were amplified via the Q5 High-Fidelity DNA Polymerase (New England Biolabs, Massachusetts, USA). Using sequence-specific primers, SacI and SacII restriction sites were respectively introduced at the 5′ and 3′ ends of both Rhopr-SKR-1 and Rhopr-SKR-2. Kozak translation initiation sequences (GCCACC) were also included at the 5′ end via the forward Rhopr-SKR primers (Table S2)55. The receptors were each cloned into the pIRES2-ZsGreen1 plasmid (Clontech, Mountain View, CA, USA).
An aequorin luminescence assay was utilized in order to measure Ca2+ signaling in HEK293/CNG cells stably expressing a modified cyclic nucleotide-gated (CNG) channel56. The cells were grown in Dulbecco’s Modified Eagle Medium Nutrient Mixture F12-Ham (DMEM/F-12) (Thermo Fisher Scientific, Waltham, MA, USA), with 1% penicillin/streptomycin, 10% fetal bovine serum (FBS), and 100 μg/mL G418 (Geneticin). The HEK293/CNG cells were consistently grown in vented T75 flasks and inclubated at 37 °C in 5% CO2. After reaching approximately 95% confluency, the mammalian cells were transiently co-transfected with the pIRES2-ZsGreen1 expression vector (containing Rhopr-SKR-1 or Rhopr-SKR-2) and the cytoplasmic luminescent reporter aequorin via X-tremeGENETM HP DNA Transfection Reagent (Roche Applied Science, Penzberg, Germany). Transfections were performed at a 2:1 ratio for the transfection reagent to the expression vectors. Cells transiently transfected with empty pIRES2-ZsGreen1 expression vectors served as a negative control. The cells were then incubated at 37 °C in 5% CO2 for 72 hours prior to performing the bioluminescence assay.
Functional receptor assays
The bioluminescence assay was performed as previously described57. At 72 hours post-transfection, the HEK293/CNG cells were harvested from the T75 flasks using a PBS with ethylenediaminetetraacetic acid (EDTA) solution then suspended in a bovine serum albumin (BSA) media (DMEM/F-12 with 1% penicillin/streptomycin and 1% BSA). Prior to the functional assay, the cells were incubated in coelenterazine h (Promega, Madison, WI, USA) to a 5 μM final concentration, and then incubated for 4 hours whilst stirring in the dark. The cells were then diluted 8-fold via BSA media. Rhopr-SK-1 and Rhopr-SK-2 (as well as nonsulfated Rhopr-SK-1) were prepared in BSA media then plated in duplicates on 96-well plates, with 50 μL of the cells then automatically injected into each of these wells via a Perkin Elmer Wallac 1420 Victor2 plate reader (Perkin Elmer, San Diego, CA, USA). The reader was used to measure luminescence response in consecutive five-second intervals (15 seconds in total). An extended Rhopr-FMRFamide (GNDNFMRFamide), tachykinin (Rhopr-TK-2), and Rhopr-Kinin-2 were used as controls to test for specificity. ATP was used as a positive control. HEK293/CNG cells transfected with empty pIRES2-ZsGreen1 vectors served as negative controls.
Second messenger pathways of Rhopr-SKRs
To test whether cAMP and/or Ca2+ are the second messengers for Rhopr-SKR-1 and Rhopr-SKR-2, HEK293/CNG cells were transfected with each receptor, as previously mentioned, then split into cells that were suspended in calcium-free DMEM with the extracellular calcium chelator EGTA (10 μM) or cells treated with the PLC inhibitor U73122 (10 μM) (Sigma Aldrich, Oakville, ON, Canada). U73122 has been previously shown to successfully inhibit the PLC pathway in the HEK293/CNG cell line56. HEK293/CNG cells with receptors that work via the Ca2+ pathway would be unable to evoke luminescence in the presence of U73122. The modified CNG channel in HEK293/CNG causes extracellular Ca2+ to flow into the cell upon an increase in cytoplasmic cAMP levels. Receptors that solely work through the cAMP pathway would be unable to produce a response in calcium-free media with EGTA. Moreover, receptors that work via both Ca2+ and cAMP would elicit a response in the presence of either U73122 or EGTA in calcium-free media.
HEK293/CNG cells were used via the same protocol as previously stated. Luminescence was measured in 5-second intervals for a total of 30 seconds. In the first set of experiments, the transfected cells were incubated in 10 μM of U73122 for 10 minutes prior to measuring luminescence in response to the addition of sulfated Rhopr-SK-1 or Rhopr-SK-2. To confirm the viability of the utilized cells, media in 1.5% Triton X-100 served as a positive control. In the second set of experiments, cells were suspended in calcium-free DMEM (Thermo Fisher Scientific, Waltham, MA, USA) with 10 μM of the extracellular calcium chelator EGTA. Here, luminescence was measured in response to sulfated Rhopr-SK-1 or Rhopr-SK-2 in calcium-free DMEM in the presence of EGTA. A batch of cells that was also pre-incubated in calcium-free DMEM with EGTA were then washed, incubated in regular DMEM, and then tested in response to sulfated Rhopr-SK-1 and Rhopr-SKR-2. Forskolin, which is a stimulator of adenylate cyclase, and the cAMP analog dibutyryl cAMP (dbcAMP) were utilized as controls to confirm the expression of functional CNG channels in the HEK293/CNG cells. ATP served as a positive control. GraphPad Prism version 8.00 (https://www.graphpad.com) was used to analyze the bioluminescence results.
Spatial expression analyses of Rhopr-SKRs
The following tissues were used from 5th instars for the spatial expression analysis: CNS, foregut, anterior midgut, posterior midgut, hindgut, salivary glands, heart, Malpighian tubules, fat body, as well as male and female reproductive tissues. RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA) was used to extract RNA from the tissues. At least 20 ng of the total RNA was used as a template for the corresponding cDNA synthesis via the iScript Reverse Transcription Supermix for RT-qPCR (BioRad Laboratories Ltd., Mississauga, ON, Canada). The RT-qPCR analyses were performed using an Mx4000 Multiplex Quantitative PCR System (Stratagene, Mississauga, ON, Canada), using SsoFast EvaGreen Supermix with Low ROX (BioRad Laboratories Ltd., Mississauga, ON, Canada). The temperature-cycling profile consisted of an initial denaturation step at 95 °C for 30 seconds, followed by 40 cycles of denaturation (5 seconds at 95 °C) and annealing/extension (24 seconds at 57 °C), which was followed by a melt curve analysis (58 °C-to-95 °C). The melt curve analysis as well as gel electrophoresis were performed to confirm the specificity of the obtained products. A 208 base-pair Rhopr-SKR-1 cDNA fragment was amplified via the forward primer, 5′ GAAGCTGGTAAAGTACAAATTC 3′, and the reverse primer, 5′ CGAAATCTTTTAGTAAGGATCCA 3′. For Rhopr-SKR-2, a 165 base-pair cDNA fragment was amplified using the forward primer, 5′ TCCAAGCTGTTTCAGTATCAGTAG 3′, and the reverse primer, 5′ GCGAATTCCACACTAAAGACA 3′. Primer efficiencies were calculated for the respective target Rhopr-SKR. Three reference genes, namely ribosomal protein 49, β-actin and α-tubulin, were also amplified via the primers previously described by Zandawala et al.57 (Table S3). The levels of expressed transcripts were quantified via the standard curve method. The delta-delta Ct (ΔΔCT) method was applied to calculate the relative expression of the transcripts. The transcript expression levels for the Rhopr-SKRs were normalized via geometric averaging of the transcript levels of the housekeeping/reference genes. Each reaction contained two technical replicates as well as a no template control and a no reverse transcriptase control. Three biological replicates were performed.
Knockdown of Rhopr-SKs and Rhopr-SKRs transcripts via double stranded RNA (dsRNA)
Rhopr-SK-specific primers containing the T7 RNA polymerase promoter sequence overhang was utilized in generating dsRNA, which was injected into 5th instar R. prolixus, thereby silencing the transcript expression of Rhopr-SK, which encodes Rhopr-SK-1 and Rhopr-SK-2. A 23 base pair T7 RNA polymerase promoter sequence overhang (5′ TAATACGACTCACTATAGGGAGA 3′) was conjugated onto the 5′ ends of forward and reverse Rhopr-SK or Rhopr-SKR gene-specific primers. Using pre-amplified Rhopr-SK and Rhopr-SKR transcripts as templates, the appropriate primers containing the T7 overhangs were utilized to create new cDNA templates (Table S4). The new cDNA templates were then utilized for the synthesis of double-stranded RNA (dsRNA) via the T7 Ribomax Express RNAi System (Promega, Madison, WI, USA). The length of the dsRNA was 207 base pairs for Rhopr-SK, 208 base pairs for Rhopr-SKR-1, and 165 base pairs for Rhopr-SKR-2. The dsRNA utilized for the knockdown of Rhopr-SKR-1 and Rhopr-SKR-2 were synthesized separately then combined pre-injection. Following the anesthetisation of R. prolixus 5th instars with CO2, 1 μg of the Rhopr-SK (or Rhopr-SKR-1 with Rhopr-SKR-2) dsRNA was injected into the thoraces of the insects. All controls were injected with 1 μg of ampicillin resistance gene dsRNA (dsARG), which targets a transcript that is not present in R. prolixus.
Prior to the feeding assays, for both of the Rhopr-SK and Rhopr-SKR knockdown experiments, RT-qPCR was performed on insects injected with the appropriate dsRNA at days 2, 4, and 6 post-injection. This was done to determine the most optimal transcript-knockdown day, during which the injected insects would be fed. For the Rhopr-SKR experiments, the primers utilized for the RT-qPCR reactions were the same as the ones previously utilized for spatial expression analyses (Table S3). Five of the injected animals were dissected per experiment, and PureLink RNA Mini Kit (Life Technologies Corporation, Carlsbad, CA, USA) was used to extract RNA from the CNS tissues. Using the extracted RNA, cDNA was synthesized via the iScript Reverse Transcription Supermix (BioRad Laboratories Ltd., Mississauga, ON, Canada). RT-qPCR was performed using the synthesized CNS cDNA as a template, with an annealing temperature of 60 °C, and the same settings as previously described. For both Rhopr-SKs and Rhopr-SKRs, RT-qPCR results revealed that the knockdown of the transcripts was most effective two days post-injection. RT-qPCR was also performed on a subset of the injected insects (5 insects per experiment) on the day of feeding (two days post-injection) to confirm the efficiency of the injected dsRNA on the knockdown of the respective transcripts. Each reaction contained two technical replicates as well as a no template control; three biological replicates were performed.
Two days following the injection, feeding was examined. The animals were weighed prior to feeding, then allowed access to 37 °C defibrinated rabbit blood (Cedarlane Laboratories Inc., Burlington, ON, Canada) for 20 minutes. Following the feeding period, the animals were individually weighed. The animals were then weighed 30 minutes post-feeding, every hour for the first 5 hours post-feeding, and then 24 hours post-feeding, in order to monitor excretion.
For experiments using dsRNA for Rhopr-SKs, immunohistochemistry was performed to visually confirm the knockdown of the Rhopr-SK transcript (and therefore absence of expressed SKs) in CNS tissues. The tissues were dissected from two days post-injection with Rhopr-SK dsRNA, and immunohistochemistry was performed as previously described17. Immunohistochemistry was also performed on control insects injected with dsARG.
Statistical analyses
GraphPad Prism version 8.00 (https://www.graphpad.com) was used to statistically analyse the data. All of the relevant data is available upon request.
References
Rehfeld, J. F. Cholecystokinin – From local gut hormone to ubiquitous messenger. Front. Endocrinol. 8, 47 (2017).
Nachman, R. J., Holman, G. M., Cook, B. J., Haddon, W. F. & Ling, N. Leucosulfakinin-II, a blocked sulfated insect neuropeptide with homology to cholecystokinin and gastrin. Biochem. Biophys. Res. Comm. 140, 357–364 (1986).
Nachman, R. J., Holman, G. M., Haddon, W. F. & Ling, N. Leucosulfakinin, a sulfated insect neuropeptide with homology to gastrin and cholecystokinin. Science 234, 71–73 (1986).
Schoofs, L., Janssen, T. & Nachman, R. J. Sulfakinins in Handbook of Biol. Act. Peptides (ed. Kastin, A. J.) 310–314 (Academic Press, 2013).
Kollien, A. & Schaub, G. The development of Trypanosoma cruzi in triatominae. Parasitol. Today 16(9), 381–387 (2000).
Buxton, P. A. The biology of a blood-sucking bug, Rhodnius prolixus. Trans. Roy. Entomol. Soc. Lond. 78(2), 227–256 (1930).
Verleyen, P. et al. Neuropeptidomics of the grey flesh fly, Neobellieria bullata. Biochem. Biophys. Res. Comm. 316, 763–770 (2004).
Li, B. et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Gen. Res. 18, 113–122 (2008).
Ons, S., Richter, F., Urlaub, H. & Pomar, R. R. The neuropeptidome of Rhodnius prolixus. brain. Proteom. 9, 788–792 (2009).
Clynen, E. & Schoofs, L. Peptidomic survey of the locust neuroendocrine system. Insect Biochem. Mol. Biol. 39, 491–507 (2009).
Hauser, F. et al. Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. J. Proteom. Res. 9, 5296–5310 (2010).
Huybrechts, J. et al. Neuropeptide and neurohormone precursors in the pea aphid, Acyrthosiphon pisum. Ins. Mol. Biol. 19, 87–95 (2010).
Audsley, N., Matthews, H. J., Down, R. E. & Weaver, R. J. Neuropeptides associated with the central nervous system of the cabbage root fly. Delia radicum (L.) Peptides 32, 434–440 (2011).
Zoephel, J., Reiher, W., Rexer, K. H., Kahnt, J. & Wegener, C. Peptidomics of the Agriculturally Damaging Larval Stage of the Cabbage Root Fly Delia radicum (Diptera: Anthomyiidae). Plos One 7, e41543 (2012).
Nichols, R., Schneuwly, S. A. & Dixon, J. E. Identification and characterization of a Drosophila homologue to the vertebrate neuropeptide cholecystokinin. J. Biol. Chem. 263, 12167–12170 (1988).
Kubiak, T. M. et al. Cloning and functional expression of the first Drosophila melanogaster sulfakinin receptor DSK-R1. Biochem. Bioph. Res. Comm. 291, 313–320 (2002).
Al-Alkawi, H., Lange, A. B. & Orchard, I. Cloning, localization, and physiological effects of sulfakinin in the kissing bug, Rhodnius prolixus. Peptides 98, 15–22 (2017).
Veenstra, J. A. Isolation and structure of two gastrin/CCK-like neuropeptides from the American cockroach homologous to the leucosulfakinins. Neuropep. 14, 145–149 (1989).
Fonagy, A., Schoofs, L., Proost, P., Van Damme, J. & De Loof, A. Isolation and primary structure of two sulfakinin-like peptides from the fleshfly, Neobellieria bullata. Comp. Biochem. Phys. 103, 135–142 (1992).
Maestro, J. L. et al. Screening of antifeedant activity in brain extracts led to the identification of sulfakinin as a satiety promoter in the German cockroach. Euro. J. Biochem. 268, 5824–5830 (2001).
Nichols, R. The first nonsulfated sulfakinin activity reported suggests nsDSK acts in gut biology. Peptides 28(4), 767–773 (2007).
Palmer, G. C., Tran, T., Duttlinger, A. & Nichols, R. The drosulfakinin 0 (DSK 0) peptide encoded in the conserved Dsk gene affects adult Drosophila melanogaster crop contractions. J. Ins. Phys. 53, 1125–1133 (2007).
Nichols, R., Manoogian, B., Walling, E. & Mispelon, M. Plasticity in the effects of sulfated and nonsulfated sulfakinin on heart contractions. Front. Biosci. 14, 4035–4043 (2009).
Marciniak, P., Kuczer, M. & Rosinski, G. New physiological activities of myosuppressin, sulfakinin and NVP-like peptide in Zophobas atratus beetle. J. Comp. Phys. B. 181(6), 721–730 (2011).
Yu, N., Nachman, R. J. & Smagghe, G. Characterization of sulfakinin and sulfakinin receptor and their roles in food intake in the red flour beetle Tribolium castaneum. Gen. Comp. Endocrin. 188, 196–203 (2013).
Downer, K. E., Haselton, A. T., Nachman, R. J. & Stoffolano, J. G. Jr. Insect satiety: sulfakinin localization and the effect of drosulfakinin on protein and carbohydrate ingestion in the blow fly, Phormia regina (Diptera: Calliphoridae). J. Ins. Phys. 53, 106–112 (2007).
Wei., Z. et al. Sulfakinins reduce food intake in the desert locust, Schistocerca gregaria. J. Ins. Phys. 46, 1259–1265 (2000).
Meyering-Vos, M. & Müller, A. RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J. Ins. Phys. 53, 840–848 (2007).
Yu, N., Swevers, L., Nachman, R. J. & Smagghe, G. Development of cell-based bioassay with Sf9 cells expressing TcSKR1 and TcSKR2 and differential activation by sulfated and non-sulfated SK peptides. Peptides 53, 238–242 (2014).
Dufresne, M., Seva, C. & Fourmy, D. Cholecystokinin and gastrin receptors. Phys. Rev. 86, 805–847 (2006).
Predel, R. et al. Posttranslational modifications of the insect sulfakinins: sulfation, pyroglutamateformation and O-methylation of glutamic acid. Euro. J. Biochem. 263, 552–560 (1999).
Brody, T. & Cravchik, A. Drosophila melanogaster G protein-coupled receptors. J. Cell Biol. 150(2), F83–F88 (2000).
Hewes, R. S. & Taghert, P. H. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster. genome. Gen. Res. 11(6), 1126–1142 (2001).
Wicher, D., Derst, C., Gautier, H., Lapied, B. & Heinemann, S. H. The satiety signaling neuropeptide perisulfakinin inhibits the activity of central neurons promoting general activity. Front. Cell. Neurosci. 1, 3 (2007).
Chen, X. & Ganetzky, B. A neuropeptide signaling pathway regulates synaptic growth in Drosophila. J. Cell Biol. 196(4), 529–543 (2012).
Zels, S. et al. Signaling properties and pharmacological analysis of two sulfakinin receptors from the red flour beetle, Tribolium castaneum. PloS One 9(4), e94502 (2014).
Duve, H., Rehfeld, J. F., East, P. & Thorpe, A. Localisation of sulfakinin neuronal pathways in the blowfly Calliphora vomitoria. Cell Tiss. Res. 275, 177–186 (1994).
Piiper, A., Stryjek-Kaminska, D., Klengel, R. & Zeuzem, S. CCK, carbachol, and bombesin activate distinct PLC-beta isoenzymes via Gq/11 in rat pancreatic acinar membranes. Amer. J. Phys. – Gastroin. Liv. Phys. 272(1), 135–140 (1997).
Yule, D. I., Baker, C. W. & Williams, J. A. Calcium signaling in rat pancreatic acinar cells: a role for Gαq, Gα11, and Gα14. Amer. J. Phys. – Gastroin. Liv. Phys. 276, 271–279 (1999).
Paulssen, R. H., Fraeyman, N. & Florholmen, J. Activation of phospholipase C by cholecystokinin receptor subtypes with different G-protein-coupling specificities in hormone-secreting pancreatic cell lines. Biochem. Pharmacol. 60(6), 865–875 (2000).
Janssen, T. et al. Discovery of a cholecystokinin-gastrin-like signaling system in nematodes. Endocrin. 149(6), 2826–2839 (2008).
Staljanssens, D. et al. The CCK (−like) receptor in the animal kingdom: functions, evolution and structures. Peptides 32(3), 607–619 (2011).
Pisegna, J. R., de Weerth, A., Huppi, K. & Wank, S. A. Molecular cloning of the human brain and gastric cholecystokinin receptor: structure, functional expression and chromosomal localization. Biochem. Biophys. Res. Comm. 189(1), 296–303 (1992).
Langhans, N. et al. Abnormal gastric histology and decreased acid production in cholecystokinin-B/gastrin receptor-deficient mice. Gastroenter. 112(1), 280–286 (1997).
Chen, X., Peterson, J., Nachman, R. J. & Ganetzky, B. Drosulfakinin activates CCKLR-17D1 and promotes larval locomotion and escape response in Drosophila. Fly 6, 290–297 (2012).
Harshini, S., Nachman, R. J. & Sreekumar, S. In vitro release of digestive enzymes by FMRF amide related neuropeptides and analogues in the lepidopteran insect Opisina arenosella (Walk.). Peptides 23(10), 1759–1763 (2002).
Nichols, R., Egle, J. P., Langan, N. R. & Palmer, G. C. The different effects of structurally related sulfakinins on Drosophila melanogaster odor preference and locomotion suggest involvement of distinct mechanisms. Peptides 29(12), 2128–2135 (2008).
Nachman, R. J. et al. Insect myosuppressins and sulfakinins stimulate release of the digestive enzyme α-amylase in two invertebrates: the scallop Pecten maximus and insect Rhynchophorus ferrugineus. Ann. New York Acad. Sci. 814(1), 335–338 (1997).
Kiss, B. et al. Selective elimination/RNAi silencing of FMRF-related peptides and their receptors decreases the locomotor activity in Drosophila melanogaster. Gen. Comp. Endocrin. 191, 137–145 (2013).
Slocinska, M., Marciniak, P., Jarmuszkiewicz, W. & Rosinski, G. New metabolic activity of the nonsulfated sulfakinin Zopat-SK-1 in the insect fat body. Peptides 68, 157–163 (2015).
Veenstra, J. A., Lau, G. W. & Petzel, D. H. Immunohistological localization of regulatory peptides in the midgut of the female mosquito Aedes aegypti. Histochem. Cell Biol. 104(5), 337–347 (1995).
Ons, S. et al. Identification of G protein coupled receptors for opsines and neurohormones in Rhodnius prolixus. Genomic and transcriptomic analysis. Ins. Biochem. Mol. Bio. 69, 34–50 (2016).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Bio. 215(3), 403–410 (1990).
Paluzzi, J. P., Russell, W. K., Nachman, R. J. & Orchard, I. Isolation, cloning, and expression mapping of a gene encoding an antidiuretic hormone and other CAPA-related peptides in the disease vector, Rhodnius prolixus. Endocrin. 149(9), 4638–4646 (2008).
Kozak, M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Bio. 196(4), 947–950 (1987).
Visegrady, A., Boros, A., Némethy, Z., Kiss, B. & Keserű, G. M. Application of the BD ACT One™ Technology for the high-throughput screening of Gs-coupled receptor antagonists. J. Biomol. Scr. 12(8), 1068–1073 (2007).
Zandawala, M., Li, S., Hauser, F., Grimmelikhuijzen, C. J. P. & Orchard, I. Isolation and functional characterization of calcitonin-like diuretic hormone receptors in Rhodnius prolixus. PLoS One 8(11), e82466 (2013).
Acknowledgements
We thank Sam Hana for his guidance, Dr. Adriano Senatore for providing the pIRES2-ZsGreen1 plasmid, and Dr. Jan Veenstra for providing the SK antiserum. This research was supported through NSERC Discovery grants to A.B.L. and I.O.
Author information
Authors and Affiliations
Contributions
M.B., A.B.L. and I.O. designed the experiments and mapped out the manuscript. M.B. performed the experiments, wrote the manuscript and prepared all the figures. A.B.L. and I.O. reviewed and contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bloom, M., Lange, A.B. & Orchard, I. Identification, Functional Characterization, and Pharmacological Analysis of Two Sulfakinin Receptors in the Medically-Important Insect Rhodnius prolixus. Sci Rep 9, 13437 (2019). https://doi.org/10.1038/s41598-019-49790-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-49790-x
This article is cited by
-
Cholecystokinin/sulfakinin peptide signaling: conserved roles at the intersection between feeding, mating and aggression
Cellular and Molecular Life Sciences (2022)
-
Sulfakinins influence lipid composition and insulin-like peptides level in oenocytes of Zophobas atratus beetles
Journal of Comparative Physiology B (2022)
-
Identification of sulfakinin receptors (SKR) in Tenebrio molitor beetle and the influence of sulfakinins on carbohydrates metabolism
Journal of Comparative Physiology B (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.