Abstract
DNA purification is essential for the detection of human clinical specimens. A non-destructive, controllable, and low reagent consuming DNA extraction method is described. Negatively charged DNA is absorbed onto a negatively charged montmorillonite to achieve non-destructive DNA extraction based on cation bridge construction and electric double layer formation. Different valence cation modified montmorillonite forms were used to validate the charge-dependent nature of DNA adsorption on montmorillonite. Electric double layer thickness thinning/thickening with the high/lower valence cations exists, and the minerals tended to be sedimentation-stable due to the Van der Waals attraction/electrostatic repulsion. Li-modified montmorillonite with the lowest charge states showed the best DNA adsorption efficiency of 8–10 ng/μg. Charge-dependent regulating research provides a new perspective for controllable DNA extraction and a deep analysis of interface engineering mechanisms.
Similar content being viewed by others
Introduction
DNA purification has been widely used for transfection, sequencing, and polymerase chain reaction (PCR)1. Sample preparation steps, including adsorption and desorption of DNA are of prime importance for the following analysis procedures, using the physicochemical characteristics of the negative charge (owing to the phosphate group (PO43−))2,3 and the different solubilities in organic and aqueous phases. Traditional chemical extraction, magnetic separation4,5,6,7,8,9 and silicon material matrix separation, such as SiO210,11,12,13,14,15 and other extraction methods16,17,18,19 (the corresponded adsorption pattern is shown in Table S1), have been developed over the past decades. Generally, surface charging is the principal method currently available for strengthening the interaction between DNA and materials in the solid phase extraction process. Despite rapid advances in performance improvement of DNA extraction efficiency, less attention has been paid to the influence of the properties of modified materials at the same time. Therefore, a conceptually different method was employed for charge-dependent extraction of DNA based on the DLVO theory (a dispersion stabilizing theory).
Natural clay, as an emerging biomaterial for biomedical engineering20,21,22,23,24,25,26,27,28,29, has the advantages of stable chemical composition30,31, various morphology structures and the specific physicochemical characteristics32,33,34. Montmorillonite (MMT, Al2Si4O10(OH)4·nH2O) is a 2:1 layered clay mineral that consists of one Al-octahedral sheet sandwiched between two Si-tetrahedral sheets31,35. The natural negative charge is derived from the crystalline substitution, and the charge state could be altered by electric double layer regulation or cation exchange. In addition, the broken edges of silicate clay minerals adsorb hydrogen ions in different pH medium environments and could affect the charge. The unique structure and natural characteristics36,37 expand its applied range for biomedical engineering38,39,40,41.
Obviously, the positive charge facilitates adsorption by the strong electrostatic attraction between the opposite electricity. However, it is unclear whether the negative electricity will serve as a regulator of the adsorption process. It is easy to see that the adsorption between two negative materials by weaker electrostatic attraction, such as a cation bridge, is more conducive to non-destructive desorption. Inspired by this idea, negatively charged DNA (purified from HeLa cells) is absorbed onto the negatively charged montmorillonite to achieve a non-destructive DNA extraction based on the cation bridge construction and the electric double layer formation (Fig. 1). An ideal single factor analysis is established. The adsorption was highly correlated with charge state. The relationship between the different valence cations and the electric double layer thickness formation under the DLVO theory was clarified. The charge-dependent and surface-interfacial relationship in DNA adsorption was detailed. Further, the relationships among the charge, the dispersion, and the cation bridge for MMT interactions with DNA were analysed.
Results
Different valence cations (Li+, Na+, Mg2+, Fe3+ and Al3+) with the same mass ratio were selected to modify the Ca-MMT to the maximum adsorption capacity, and the corresponding adsorption was measured. The adsorption and desorption of DNA could be achieved by different ion-MMT combinations (Fig. 2a and Fig. S1). Based on the unchanged brightness of agarose gel electrophoresis, the adsorption efficiency of Li-MMT, Na-MMT, Ca-MMT and Mg-MMT were not changed as time increased. Interestingly, the DNA adsorption capacity of Al-MMT and Fe-MMT varied from 5 min to 2 hours, due to the sedimentation of the Al or Fe ion hydrolysis, as indicated by the different brightness of two agarose gel electrophoresis bands. To further investigate the adsorption efficiency, Nano Drop software is utilized based on the brightness of the lines showed in Fig. 2a. Na-MMT and Li-MMT exhibit high DNA binding action with an 8–10 ng/μg adsorption capacity (Fig. 2b), and Mg-MMT and Ca-MMT show only a 4–5 ng/μg adsorption capacity.
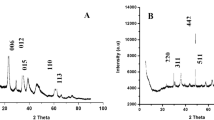
Adsorption and characterization of ion-MMT: (a) Electrophoresis analysis for the supernatant of clay binding with DNA (the intact gels of adsorption and desorption were included in a supplementary information file as shown in Fig. S1). (b) Adsorption efficiency of DNA of different samples. (c) Zeta potential of fresh samples and samples after a week. (d) XRD pattern of Na-MMT, Li-MMT, Ca-MMT, Mg-MMT, Fe-MMT and Al-MMT; the inset shows the modification efficiency of different cations measured by ICP. (e) Zeta potential of samples in different system and binding with DNA.
To further explore the possible interfacial interactions between ion-MMT and DNA, zeta potential was obtained to mirror the particles’ surface charge, and a surprising observation was made. The zeta potential of Al-MMT and Fe-MMT was changeable when the ion-MMT was remeasured two weeks later. The results showed that the zeta potential value of ion-MMT increased gradually as the cationic valence increased due to the thinner diffusion layer formed by the high cationic valence state (Fig. 2c, black line). The high valence cations would compress the thickness of the electric double layer, thus improving the zeta potential. Interestingly, the zeta potential of Al-MMT and Fe-MMT were markedly decreased from −5.76 mV and −9.85 mV to −23.7 mV and −28.2 mV after two weeks, respectively, which could be attributed to the hydrolysis of Fe3+ and Al3+ (Fig. 2c).
To determine the ions locations during the modification process, ICP and XRD were used to evaluate the ion-exchanged efficiency of MMT. XRD analysis (Figs 2d and S2) revealed that the typical diffraction pattern of Ca-MMT with a characteristic d001 value of 1.49 nm (2θ = 5.92°), shifted to 1.30 nm (7.18°), 1.29 nm (6.84°), 1.44 nm (6.12°), and 1.52 nm (5.80°), which corresponds to Na-MMT, Li-MMT, Fe-MMT and Al-MMT, respectively. The above results indicate obvious shifting with successful intercalation, slight shifting with inadequate intercalation with the most adsorbed on the external surface, and a lack of shifting, it was mainly adsorbed on the external surface (Mg-MMT). The ICP results showed that 90% of the cations were successfully modified to MMT (inset in Fig. 2d).
In addition, we found that the zeta potential of the ion-MMT in a hydrochloric acid (black) and sodium acetate (red) solution system showed different behaviours, with the exception of Fe-MMT and Al-MMT (Fig. 2e), which could be attributed to the diffusion layer thickening when the cation ions captured the negatively charged acetate groups. However, the trivalent cation ions were limited by the ion hydrolysis, and the surface cations decreased. Thus, the diffusion layer could not thicken, and the zeta potential was generally not changed. In addition, the charge state of ion-MMT@DNA remains negative and even lower than ion-MMT. Interestingly, the adsorption capacity was greatest for the lowest charge states.
The interlayer spacing of Li-MMT was decreased (Fig. 3a) and the zeta potential was negative and stable at pH >4 (Fig. 3b) when compared with the original d001 value = 1.49 nm, which was stable at approximately pH = 11 for Ca-MMT. The corresponding FTIR spectrums results (Fig. 3c and Fig. S3) showed that the O-H stretching vibration band at 3,620 cm−1 and 3,420 cm−1 correspond to the Al-OH group and the interlayer water molecule, respectively. Bending-vibration bands of O-H are observed at 1,636 cm−1 and 3,420 cm−1, which indicates that the ion-MMT interlayer contains crystal water. The stretching-vibration band of Si-O at 1,035 cm−1 and the bending-vibration band of Si-O at 465 cm−1 indicated that the layered structure of ion-MMT consisted of a silicon-oxygen tetrahedron. The bending-vibration bands of Al-OH, Mg-OH, Si-OMg and Si-OFe were located at 915, 844, 525 and 460 cm−1, respectively. For DNA obtained using phenol-chloroform extraction, the band at 551 cm−1 corresponded to the asymmetric variable angle of PO4, and the band at 695 cm−1 was ascribed to the stretching-vibration of P = S given the remaining protein during the extraction. The sharp bands at 3,510 and 3,455 cm−1 are ascribed to the anti-symmetrical and symmetrical stretching vibration of O-H, and the band at 1,649 cm−1 corresponds to the variable angle vibration of O-H for the tested DNA when dissolved in water. For the DNA@ion-MMT samples, the asymmetric variable angle of PO4 is shifted to 624 cm−1. The bending-vibration band of Si-OMg and Si-OFe were stronger and shifted to 521 and 466 cm−1 after combining with DNA. The other vibration band in the FTIR spectrum of DNA@ion-MMT was similar to that of ion-MMT.
Characterization of Li-MMT before and after the adsorption of DNA: (a) XRD pattern and (b) Zeta potential of Li-MMT and Ca-MMT. (c) FTIR spectra of DNA, Ca-MMT, Li-MMT before and after binding with DNA. (d–f) SEM images of DNA, Li-MMT and Li-MMT@DNA; (g–i) TEM images of DNA, Li-MMT and Li-MMT@DNA; (j–l) is the SEM EDX confirming the formation of DNA, Li-MMT and Li-MMT@DNA.
SEM and TEM images with the EDX were used to survey the interfacial relationships between DNA and MMT, and the findings are presented in Fig. 3. DNA has the morphology of a short stick or elliptical with an average length of 1–20 μm, which is attributed to the agglomeration of single DNA (5–6 μm in length and 2 nm in diameter) (Fig. 3d). The TEM image of DNA indicates that the white-threadlike DNA was filamentous clumped together and formed an organic-like film (Fig. 3g). The energy-dispersive X-ray (EDX) spectra demonstrated that the DNA mainly contains C and O (Fig. 3j). Li-MMT has the morphology of a typical layered structure that was 3–4 μm in width and 1–2 μm in thickness (Fig. 3e,h). After the DNA was adsorbed, a more organic-like film was wrapped on the layered structure (Fig. 3f,i), with rare white-threadlike matters on the end edge of the Li-MMT, and the carbon content was increased from 15.14 wt.% to 24.77 wt.% due to the C element in the DNA (Fig. 3k,l).
Expression of the target gene in the host cells by plasmid transfection or transformation requires an integral plasmid DNA. Thus, the pEGFP-C1 plasmid expressing green fluorescent protein was chosen to validate the integrity of the eluted DNA. The pEGFP-C1 plasmid, which underwent an adsorption and desorption procedure, was transfected to the HeLa cells (Fig. 4a and Fig. S4). The eluted plasmid expressed GFP protein in HeLa cells, and green fluorescence was observed. In addition, similar results were obtained with the intact pEGFP-C1 plasmid (pEGFP-C1 plasmid positive control). No green fluorescence was observed in the HeLa cells without plasmid transfection (negative control). The same results were obtained when we transformed E.coli (Escherichia coli) with the eluted plasmid (Fig. 4b). These results confirmed that the eluted DNA is undamaged, functional and ready to perform multiple downstream applications.
DNA eluted from ion-MMT do not effect the function of DNA (a) pEGFP-C1 plasmids maintained their expression function(green) after adsorption and desorption was transfected into HeLa. The nuclei was stained by DAPI (blue). (b) pEGFP-C1 plasmids maintained their expression function after adsorption and desorption was transformed to E. coli. The clones were counted after kanamycin selection. (c) Statistic analysis was performed for (b). ns, no significant difference.
Discussion
Dispersion effects of ion-MMT modulate the adsorption efficiency. The classical DLVO theory is currently used as a benchmark to reasonably discuss the attractive or repulsive force between surfaces of charged particles, and further describe the dispersion and sedimentation of particles. Van der Waals attraction and electrostatic repulsion are involved into the stability of the particles’ solution. Essentially, the thickness of electrical double layer is a critical factor. The thickness of the electrical double layer is known as the Debye length κ−1 based on the following expression:
where e is the electronic charge (1.062 × 10−19 C), NA is the constant of Avogadro 6.022 × 1023 mol−1, Z is the valency of the ion. Here, Li+ has a valency of +1, Ca2+ has a valency of +2 and Al3+ has a valence of +3, C is the molar concentration of ion, ε is the absolute dielectric constant of dispersive medium, k is the constant of Boltzmann 1.381 × 10−23 J, and T is the absolute temperature.
Apparently, the Debye length κ−1 is inversely proportional to the ion valence when C and ε remain the same in different samples. Suppose the charge is low, i.e., the Z value is small, the corresponding value of κ−1 is large, and the double electric layer formed is thicker. Two particles approach each other, and the interference between their electrical double layers increased and overlapped. Thus, the particle-particle electrostatic repulsion increases, and a relatively stable state was achieved. Simultaneously, when the electric double layer of the charged surface is thin, the Van der Waals attraction dominants, and the particles get closer. Thus, the particles’ solution tends to the sedimentation state (Fig. 5a).
As a result, the MMT solution’s stability depends on the different valence cations that are modified based on the electric double layer theory. Electric double layer thickness is reduced/increased with high/lower valence cations, and the minerals tended to be sedimentation- stable due to the Van der Waals attraction/electrostatic repulsion.
Further, the DNA adsorption was based on the cation bridge construction (Fig. 5b), but it was profoundly affected by the thickness of the electric double layer formation of the charged surface. The electric double layer thickness increased in the presence of lower valence cations (e.g., Li+, Na+). Minerals with lower charge states (more negative) tend to be stable due to the electrostatic repulsion (Fig. 6a); thus, the DNA could fully contact the MMT. The electric double layer thickness decreases in the presence of high valence cations (e.g., Fe3+, Al3+). Minerals with higher charge states (less negative) tend towards sedimentation due to the Van der Waals attraction (Fig. 6a); thus, only a few DNA are adsorbed on the MMT. Phenomenon of dispersion and agglomeration has an important bearing on the valence state of ions. The minimum concentration follows the Schulze-Hardy rule42:
where Z is the valency of the ion, and n is 6 in three dimensions. Therefore, the minimum concentration of monovalent, two valence and trivalent ions to trigger agglomeration is approximately 1: (1/2)6: (1/3)6. Metal cations with different valence of ion-MMT are consistent with this rule. Thus, rapid sedimentation can be achieved with low concentrations of high-valence metal cations. Generally, adsorption is difficult to occur between substances that are both negatively charged. Cations could act as a bridge and thus facilitate adsorption if it is introduced into the middle of particles. Repulsion forces will be generated between “bridges” with moderate length, which make particles simultaneously more disperse and stabile. Low-valence cations form “bridges” with moderate length (the double layer is thicker) on the surface of materials, the excellent dispersion make it possible for “bridges” to connect with DNA. The “bridge” formed by the high-valence cation on the surface of the material is shorter (the electric double layer is thinner), and the particles have been agglomerated before most of the “bridge” is attached to DNA, thereby reducing the amount of adsorption.
Most of the previous investigations believed that three adsorption modes exist: electrostatic interaction, cation bridge and ligand exchange. The electrostatic interaction is the dominant relaxation mechanism when the nanomaterial surface is positively charged. The cation bridge plays a major role when both the material surface and DNA are negatively charged. The ligand exchange is typically used to explain the interaction between the nanomaterial surface (with hydroxyl groups) and DNA (with the phosphate groups). In contrast to the existing literature reports, this paper proposed a creative DNA adsorption based on the electric double layer theory for material design and the cation bridge mechanism for interface analysis. In addition, the surface and interface analysis by the atom force microscope (AFM) illustrate that the roughness of the MMT surface was significantly reduced after DNA adsorption (Fig. 6b,c).
Natural two-dimensional (2D) MMT nanoclay has been incorporated into a DNA purification system. The interfacial interaction between ion-MMT and DNA was studied while maintaining the overall negative charge. The adsorption and desorption of DNA could be achieved by different ion-MMT combinations. The expression of green fluorescent protein from the eluted DNA indicates its excellent integrity. Based on the aforementioned results, we demonstrate a strong correlation between the DNA adsorption efficiency and the electric double layer construction. The DNA adsorption mechanism involves cation bridge construction and electric double layer formation which could be regulated by the surface charge. The degree of combination between DNA and ion-MMT was also strongly charge dependent. The lowest charge states (more negative) exhibit high adsorption behaviour with a maximum adsorption. Due to the electrostatic repulsion between particles with a thicker electric double layer, the lowest charge states make the system more dispersed and stable. As the charge state increases, the accelerated sedimentation of particles become stronger, due to the Van der Waals attraction of particles with thinner electric double layer. The electric double layer provides the desired cation for the cationic bridge to produce DNA adsorption. However, the dispersion and sedimentation of ion-MMT caused by the thickness of the electric double layer are more closely related to the adsorption efficiency. This study provides a detailed explanation for DNA adsorbing to MMT, which focuses on the in-depth analysis of the interfacial interaction during the non-destructive purification.
Methods
Materials
MMT obtained from Zhejiang, China was purified to produce ultrafine clay powders (>99% Ca-MMT). All chemicals were analytical-reagent grade. MgCl2·6H2O was purchased from Xilong Chemical Reagent Co., Ltd. The other chemicals, including Na2CO3, Li2CO3, and Fe(NO3)3·9H2O, Al(NO3)3·9H2O were purchased from Sinopharm Chemical Reagent Co., Ltd.
Cationic modified montmorillonite
Briefly, 0.47 g of inorganic salt, including Na2CO3, Li2CO3, MgCl2·6H2O, Fe(NO3)3·9H2O, Al(NO3)3·9H2O and 1.00 g Ca-MMT, were respectively added to 9 mL of deionized water (including two aqueous solutions at pH 2 for trivalent ions), and the mixture suspension was mechanically stirred for 24 h at 60 °C. Among them, Ca-MMT was first added into the water prior to Li2CO3 for Li-MMT. Following this, the cationic modified MMT was precipitated by centrifugation. The final products were obtained after drying at 60 °C for 24 h.
Characterization
Powder X-ray diffraction (XRD, RIGAKU D/max-2550 PC) measurements of the samples were performed with a DX-2700 X-ray diffractometer using Cu Kα radiation (λ = 0.15406 nm) at a scan rate of 0.02°/s. The Fourier transform infrared (FTIR) spectra of the samples were obtained using an IRAffinity-1 FTIR spectrophotometer with KBr pellets. Scanning electron microscopy (SEM) was performed using a TESCAN MIRA3 LMU scanning electron microscope at an accelerating voltage at 20 kV, which was equipped with an Oxford X-Max20 energy spectrum system and Gatan H1002 hot table system.
Fragmented DNA preparation
Genomic DNA was purified from HeLa cells using phenol-chloroform extraction. Approximately 1.0 × 107 HeLa cells were digested from 10 cm culture dish with 0.05% trypsin and washed twice in PBS. After washing, the cells were treated with proteinase K in 3 mL lysis buffer containing 2% SDS and 10 mM Tris-HCl, pH 8.0 overnight. The digestion mixture was extracted sequentially with 6 mL phenol and 6 mL chloroform. DNA was precipitated by adding the supernatant to 10 mL ethanol. The DNA precipitation was washed twice with 500 μL 70% ethanol and diluted in 1 mL double distilled water. The diluted DNA was then fragmented by 10 s sonication. Concentration of the fragmented DNA was detected using a spectrophotometer based on 260 nm absorbance. Then, the fragmented DNA was diluted to 200 ng/μL with double distilled water.
Adsorption and desorption of DNA
Briefly, 30 mg of clay (Ca-MMT or modified MMT) was first added to 1 mL of double distilled water to make clay suspension. Then, 10 μL of MMT suspension (containing 300 μg of MMT) was then mixed with 50 μL fragmented DNA (containing 10 μg of DNA), 5 μL of 3 M NaAc (pH 5.0) and 35 μL double distilled water to make a 100 μL mixture. The mixture was gently shaken at RT for 5 min and centrifuged at 10,000 g for 10 min. DNA in the supernatant was spectrophotometrically accessed at 260 nm. The amount of DNA adsorbed was calculated by the difference between the amount of DNA added and that remaining in the supernatant.
The adsorption mixture described above was centrifuged at 10,000 g for 10 min. The residue was then washed with 100 μL double distilled water once followed by 100 μL TE buffer (containing 10 mM Tris, pH 8.0 and 1 mM EDTA) to elute the adsorbed DNA.
A similar adsorption and desorption procedure was performed with 10 μg pEGFP-C1 plasmid for further transfection experiments.
Cell culture and transfection
HeLa cells were grown in DMEM/high glucose containing 10% FBS in 5% CO2. Before transfection, cells were seeded onto glass coverslips in 24-well plates, and plasmid transfection was performed with Lipofectamine 3000 reagent (Invitrogen, San Diego, USA) according to manufacturer’s protocol. Then, 500 ng pEGFP-C1 plasmid pre- or post-material adsorption and desorption were transfected to a unique well, and an equal quantity of intact pcDNA3.1 plasmid was used for negative control cells. After 48 hours, cells were fixed for immunofluorescence.
Immunofluorescence
Forty-eight hours after transfection, HeLa cells were washed twice with PBS, fixed with paraformaldehyde (Sigma-Aldrich, St. Louis, USA) (4% w/v) for 15 min, and permeabilized with 0.1% PBST (Triton X-100 in PBS) for 5 min. The coverslips were then incubated with 4′,6-diamidino-2-phenylindole(DAPI; Sigma-Aldrich, St.Louis, USA) for 5 min to stain cell nuclei. The cells were then mounted with ProLong Gold Antifade mountant (Thermo Fisher Scientific, Waltham, USA), and observed using laser-scanning confocal microscope (TCS SP5, Leica, Wetzlar, Germany).
References
Zhu, J. Y. et al. Acidity-responsive gene delivery for “superfast” nuclear translocation and transfection with high efficiency. Biomaterials 83, 79–92 (2016).
Jain, M. L., Bruice, P. Y., Szabó, I. E. & Bruice, T. C. Incorporation of positively charged linkages into DNA and RNA backbones: a novel strategy for antigene and antisense Agents. Chem. Rev. 112, 1284–1309 (2012).
Schmidtgall, B., Spork, A. P., Wachowius, F., Höbartner, C. & Ducho, C. Synthesis and properties of DNA oligonucleotides with a zwitterionic backbone structure. Chem. Commun. 50, 13742–13745 (2014).
Tanaka, T., Sakai, R., Kobayashi, R., Hatakeyama, K. & Matsunaga, T. Contributions of phosphate to DNA adsorption/desorption behaviors on aminosilane-modified magnetic nanoparticles. Langmuir 25, 2956–2961 (2009).
Kang, K. et al. Preparation and characterization of chemically functionalized silica-coated magnetic nanoparticles as a DNA separator. J. Phys. Chem. B 113, 536–543 (2009).
Probst, C. E., Zrazhevskiy, P. & Gao, X. Rapid multitarget immunomagnetic separation through programmable DNA linker displacement. J. Am. Chem. Soc. 133, 17126–17129 (2011).
Doyle, P. S., Bibette, J., Bancaud, A. & Viovy, J. L. Self-assembled magnetic matrices for DNA separation chips. Science 295, 2237 (2022).
Liu, B. & Liu, J. DNA Adsorption by magnetic iron oxide nanoparticles and its application for arsenate detection. Chem. Commun. 50, 8568–8570 (2014).
Liu, J. et al. Magnetic silica spheres with large nanopores for nucleic acid adsorption and cellular uptake. Biomaterials 33, 970–978 (2012).
Vandeventer, P. E., Mejia, J., Nadim, A., Johal, M. S. & Niemz, A. DNA adsorption to and elution from silica surfaces: influence of amino acid buffers. J. Phys. Chem. B 117, 10742–10749 (2013).
Reinhardt, N., Adumeau, L., Lambert, O., Ravaine, S. & Mornet, S. Quaternary ammonium groups exposed at the surface of silica nanoparticles suitable for DNA complexation in the presence of cationic lipids. J. Phys. Chem. B 119, 6401–6411 (2015).
Vandeventer, P. E. et al. Multiphasic DNA adsorption to silica surfaces under varying buffer, pH, and ionic strength conditions. J. Phys. Chem. B 116, 5661–5670 (2012).
Li, X., Zhang, J. & Gu, H. Study on the adsorption mechanism of DNA with mesoporous silica nanoparticles in aqueous solution. Langmuir 28, 2827–2834 (2012).
Michaels, P. et al. A robust DNA interface on a silicon electrode. Chem. Commun. 50, 7878–7880 (2014).
Zhang, Y. et al. A simple thermoplastic substrate containing hierarchical silica lamellae for high-molecular-weight DNA extraction. Adv. Mater. 28, 10630–10636 (2016).
Andrea, K. A. et al. Adsorption of Oligo-DNA on magnesium aluminum layered double hydroxide nanoparticle surfaces: mechanistic implication to gene delivery. Langmuir 33, 3926–3933 (2017).
Xu, L., Feng, L., Hao, J. & Dong, S. Controlling the capture and release of DNA with a dual-responsive cationic surfactant. ACS Appl Mater Interfaces 7, 8876–8885 (2015).
Reinholt, S. J. & Baeumner, A. J. Microfluidic isolation of nucleic acids. Angew Chem Int Ed 53, 13988–14001 (2014).
Zhang, Y., Tang, A., Yang, H. & Ouyang, J. Applications and interfaces of halloysite nanocomposites. Appl. Clay Sci. 119, 8–17 (2016).
Dawson, J. I. & Oreffo, R. O. Clay: new opportunities for tissue regeneration and biomaterial design. Adv. Mater. 25, 4069–4086 (2013).
Liu, F. et al. Smart H2O2-responsive drug delivery system made by halloysite nanotubes and carbohydrate polymers. ACS Appl. Mater. Interfaces 9, 31626–31633 (2017).
Min, J., Braatz, R. D. & Hammond, P. T. Tunable staged release of therapeutics from layer-by-layer coatings with clay interlayer barrier. Biomaterials 35, 2507–2517 (2014).
Shi, K. et al. Novel biocompatible thermoresponsive poly(n-vinyl caprolactam)/clay nanocomposite hydrogels with macro-porous structure and improved mechanical characteristics. ACS Appl. Mater. Interfaces 9, 21979–21990 (2017).
Chimene, D., Alge, D. L. & Gaharwar, A. K. Two-dimensional nanomaterials for biomedical applications: emerging trends and future prospects. Adv. Mater. 27, 7261–7284 (2015).
Yang, J. et al. Enhanced therapeutic efficacy of doxorubicin for breast cancer using chitosan oligosaccharide modified halloysite nanotubes. ACS Appl. Mater. Interfaces 8, 26578–26590 (2016).
Long, M. et al. Emerging nanoclay composite for effective hemostasis. Adv. Funct. Mater. 28, 1704452-1–9 (2018).
Xue, J. et al. Electrospun microfiber membranes embedded with drug-loaded clay nanotubes for sustained antimicrobial protection. ACS Nano 9, 1600–1612 (2015).
Long, M. et al. Fe2O3 nanoparticles anchored on 2D kaolinite with enhanced antibacterial activity. Chem. Commun. 53, 6255–6258 (2017).
Suter, J. L., Groen, D. & Coveney, P. V. Mechanism of exfoliation and prediction of materials properties of clay-polymer nanocomposites from multiscale modeling. Nano Lett. 15, 8108–8113 (2015).
Mousa, M., Evans, N. D., Oreffo, R. O. C. & Dawson, J. I. Clay nanoparticles for regenerative medicine and biomaterial design: a review of clay bioactivity. Biomaterials 159, 204–214 (2018).
Peng, K., Fu, L., Ouyang, J. & Yang, H. Emerging parallel dual 2D composites: natural clay mineral hybridizing MoS2 and interfacial structure. Adv. Funct. Mater. 26, 2666–2675 (2016).
Peng, K., Fu, L., Yang, H., Ouyang, J. & Tang, A. Hierarchical MoS2 intercalated clay hybrid nanosheets with enhanced catalytic activity. Nano Res. 10, 570–583 (2017).
Ryu, J., Hong, D., Choi, S. & Park, S. Synthesis of ultrathin Si nanosheets from natural clays for lithium-ion battery anodes. ACS Nano 10, 2843–2851 (2016).
Zhang, Y. et al. Intercalated 2D nanoclay for emerging drug delivery in cancer therapy. Nano Res. 10, 2633–2643 (2017).
Underwood, T., Erastova, V., Cubillas, P. & Greenwell, C. H. Molecular dynamic simulations of montmorillonite-organic interactions under varying salinity: an insight into enhanced oil recovery. J. Phys. Chem. C 119, 7282–7294 (2015).
Dalmoro, V., Santos, J. H. Z. D., Armelin, E., Alemán, C. & Azambuja, D. S. Sol-gel hybrid films based on organosilane and montmorillonite for corrosion inhibition of AA2024. J. Colloid. Interface. Sci. 426, 308–313 (2014).
Carosio, F., Di, P. A., Alongi, J., Fina, A. & Saracco, G. Controlling the melt dripping of polyester fabrics by tuning the ionic strength of polyhedral oligomeric silsesquioxane and sodium montmorillonite coatings assembled through layer by layer. J. Colloid Interface Sci. 510, 142–151 (2018).
Darder, M., Aranda, P. & Ruiz-Hitzky, E. Bionanocomposites: a new concept of ecological, bioinspired, and functional hybrid materials. Adv. Mater. 19, 1309–1319 (2007).
Swadling, J. B., Coveney, P. V. & Greenwell, H. C. Clay Minerals mediate folding and regioselective interactions of RNA: a large-scale atomistic simulation study. J. Am. Chem. Soc. 132, 13750–13764 (2010).
Li, G. et al. Graphene-montmorillonite composite sponge for safe and effective hemostasis. ACS Appl. Mater. Interfaces 8, 35071–35080 (2016).
Jin, X. et al. Multifunctional cationic polymer decorated and drug intercalated layered silicate (NLS) for early gastric cancer prevention. Biomaterials 35, 3298–3308 (2014).
Guo, Y. et al. Hemin-graphene hybrid nanosheets with intrinsic peroxidase-like activity for label-free colorimetric detection of single-nucleotide polymorphism. ACS Nano 5, 1282–1290 (2011).
Acknowledgements
This work was supported by the NSFC (21878341, 51804343, 41572036 and 51225403), the Strategic Priority Research Program of Central South University (ZLXD2017005), the Natural Science Foundation of Hunan Province (2018JJ3670), the Key R&D Program of Hunan Province(2017GK2251), Hunan Provincial Science and Technology Project (2016RS2004 and 2015TP1006). The Fundamental Research Funds for the Central Universities of Central South University (2017zzts572).
Author information
Authors and Affiliations
Contributions
Y.Z. and H.Y. developed the concept. Y.Z., H.Y. and J.T. conceived the project and designed the experiments. Y.Z. and S.L. wrote the final paper. Y.Z. and H.X. wrote initial drafts of the manuscript. H.X. and Z.W. performed the experiment and data analysis. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xie, H., Wan, Z., Liu, S. et al. Charge-Dependent Regulation in DNA Adsorption on 2D Clay Minerals. Sci Rep 9, 6808 (2019). https://doi.org/10.1038/s41598-019-41093-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41093-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.