Abstract
Dentin sialoprotein (DSP) is a dentin extracellular matrix protein. It is involved in dental mesenchymal cell lineages and dentin formation through regulation of its target gene expression. DSP mutations cause dentin genetic diseases. However, mechanisms of DSP in controlling dental mesenchymal cell differentiation are unknown. Using DSP as bait, we screened a protein library from mouse odontoblastic cells and found that DSP is a ligand and binds to cell surface receptor, occludin. Further study identified that the C-terminal DSP domainaa 363–458 interacts with the occludin extracellular loop 2aa 194–241. The C-terminal DSP domain induced phosphorylation of occludin Ser490 and focal adhesion kinase (FAK) Ser722 and Tyr576. Coexpression of DSP, occludin and FAK was detected in dental mesenchymal cells during tooth development. Occludin physically interacts with FAK, and occludin and FAK phosphorylation can be blocked by DSP and occludin antibodies. This DSP domain facilitates dental mesenchymal cell differentiation and mineralization. Furthermore, transplantation and pulp-capping procedures revealed that this DSP domain induces endogenous dental pulp mesenchymal cell proliferation, differentiation and migration, while stimulating blood vessel proliferation. This study elucidates the mechanism of DSP in dental mesenchymal lineages and implies that DSP may serve as a therapeutic agent for dentin-pulp complex regeneration in dental caries.
Similar content being viewed by others
Introduction
Craniofacial skeleton is original from neural crest-derived mesenchymal cells1. These cells proliferate and differentiate into odontoblasts and osteoblasts as well as finally build dynamic mineralized tissues such as bone and dentin. In this process, cell proliferation and differentiation are tightly controlled by spatiotemporal cell-cell interaction and extracellular matrix (ECM) to ensure that the tissue attains specific size, shape, structure, and function.
ECM often provides specific microenvironments (niches) necessary for controlling morphology, cell fate specification, cell migration and tissue repair2. Degradation or activation of ECM proteins by proteolysis during growth, morphology and tissue repair can mediate rapid and irreversible responses to changes in the cellular niches and cell homeostasis3. ECM in bone and dentin mainly comprises a number of collagenous and non-collagenous proteins (NCPs). Among the NCPs, a family of small integrin-binding ligand N-linked glycoproteins (SIBLINGs) comprises bone sialoprotein (BSP), dentin matrix protein 1 (DMP1) and dentin sialophosphoprotein (DSPP), matrix extracellular phosphoglycoprotein (MEPE) and osteopontin (OPN). These SIBLING genes are highly expressed in mineralizing tissues related to tooth and bone development and believed to be responsible for initiating and modulating cell differentiation and mineralization processes via matrix-cell interaction. For instance, an Arg-Gly-Asp (RGD) triple peptide within several NCPs regulates intracellular signal pathways via cell membrane receptors such as integrin4. Despite their common origin, dentin and bone are dramatically different from their morphologies and physical functions. One of great differences is DSPP in the two tissues. Spatial and temporal expression of DSPP is largely restricted to odontoblasts and dentin5, 6. Expression of DSPP in odontoblasts and dentin is approximately 400 fold higher than that of osteoblasts and bone7. Although DSPP is transcribed from a single gene8, 9, full length of DSPP protein has scarcely been isolated from cells or tissues10, 11, whereas its cleavage products, dentin sialoprotein (DSP) and dentin phosphoprotein (DPP), are most abundant NCPs in odontoblasts and dentin12. DSP is further processed into small molecular fragments11, 13,14,15. Cleaved DSP fragments segregate into specific compartments within odontoblasts and dentin14, 16.
DSP and DPP play unique biological functions during tooth development17, 18. Mutations of either DSP or DPP domain in humans caused dentinogenesis imperfecta (DGI) type II (DGI-II, OMIM #125490) and type III (DGI-III, OMIM 125500) and dentin dysplasia (DD) type II (DD-II, OMIM 125420)19,20,21, the most common dentin genetic diseases. Mouse DSPP knock-out exhibited similar phenotype to that of DSPP gene mutations in human22. DPP contains an RGD domain, acting as a ligand, and binds to integrin as well as triggers intracellular signals via DPP-RGD/integrin-αvβ3 interactions23, 24. By contrast, DSP lacks a RGD domain9, and many DSPP gene mutations occur in DSP region19, 20, 25. DSP and peptides derived from DSP are able to regulate gene expression and protein phosphorylation as well as induce dental primary/stem cell differentiation9, 16, 26. Recently, we have identified that 36 amino acids of DSP domainaa 183–219 bind to integrin β6 and the DSP-integrin β6 complex stimulated phosphorylation of Smad1/5/8 proteins through p38 and Erk 1/2 protein kinases. The phosphorylated Smad1/5/8 proteins were translocalized into nuclei and bind to DSPP gene promoter, activating expression of DSPP and DMP1 genes and inducing dental mesenchymal cell differentiation and biomineralization9. However, the molecular mechanisms of DSP controlling gene expression and cell differentiation have not been completely understood.
Occludin (Ocln) is an integral membrane protein associated with the tight junctions (TJs) of cells and mainly comprises four transmembrane domains, NH2- and COOH-terminal cytoplasmic regions and two extracellular loops27, 28. The COOH-terminal domain is rich in serine, threonine and tyrosine residues, which are frequently phosphorylated by various protein kinases29. The extracellular loops of Ocln interact with a variety of cellular signaling molecules and are dynamically involved in intracellular signal transductions including protein phosphorylation/dephosphorylation and ion flux28, 30, 31. The cytoplasmic tail of Ocln is necessary for binding to its partners32. Ocln mutations in humans are involved in the pathogenesis of malformations of cortical development with band-like brain calcification and chronic kidney dysfunction15, 33,34,35. Ocln deficient mice developed deafness with dislocalization of tricellulin in cochlea36 and displayed other signs of pathological disorders such as growth retardation, dysfunction of the salivary gland, calcification in the brain and thinning of the compact bone37. Although Ocln is one of the TJ proteins, unexpectedly, Ocln knock-out mice exhibited the normal TJ strand formation, and visceral endoderm cells originating from Ocln-deficient embryonic stem cells have well-developed networks of tight-junction strands37, 38. It suggested that Ocln is also important for other biological functions. However, whether the signaling pathways by which DSP regulates intracellular activity via Ocln is not clearly understood.
Focal adhesion kinase (FAK or Ptk2) is the non-receptor bound tyrosine kinase and is involved in mediating both integrin and Ocln signaling for regulating intracellular transduction with the ECM on the outside of the cells39, 40. Phosphorylation events occurring within focal adhesions influence numerous processes including cell adhesion, shape, motility as well as cell differentiation and tissue development41, 42. FAK knockout studies showed an early embryonic lethal phenotype with extensive mesodermal deficiency and Ptk2−/− embryonic fibroblasts from these mice exhibited profound defects in migration43.
Our hypothesis is that DSP regulates intracellular signal transductions and dental mesenchymal cell differentiations via matrix-cell surface interaction. Here, we found that DSP as a ligand is capable of binding to its cell surface receptor, Ocln. Further researches revealed that the COOH-terminal DSP domain interacts with the extracellular loop 2 of Ocln. This peptide phosphorylates Ocln and FAK. Immunohistochemistry showed that the spatial-temporal distribution of DSP, Ocln and FAK is co-localized in odontoblasts during dentinogenesis. The DSP domain was capable of inducing differentiation and mineralization of dental pulp stem cells. Furthermore, this peptide induced endogenous mouse dental pulp cell proliferation and differentiation and enhanced biomineralization when this DSP peptide was implanted into mouse dental pulp chambers.
Results
DSP as a ligand binds to its cell surface receptor, Ocln
To identify whether DSP interacts with other proteins, we generated GST-DSP fusion protein (Fig. 1A,B), and cell lysis was isolated from mouse odontoblast-like cells. The DSP fusion protein was used as bait to screen the protein library. This result showed that four proteins among 110 candidates interact with DSP, including integrin β6, CD105 (endoglin), collagen IV and Ocln (Fig. 1C, Table 1. Supplementary Table 1). To further identify which domain of DSP binds to Ocln, we generated different DSP and Ocln gene constructs for protein-protein interactions. These results showed that the COOH-terminal DSP domainaa 363–458 (DSPf5) interacts with the extracellular loop 2 of Oclnaa 194–241 (OclnL2) (Fig. 1E–H). To further verify specific and saturable interactions and to evaluate the binding affinity between DSPf5 and OclnL2, we used a concentration range of DSPf5 and OclnL2 as analyzed by biotin-labeled proteins. These results manifested that the binding of DSPf5 to OclnL2 was dose- and time-dependent manners (Fig. 1I). For in vivo study, mammalian expression of DSP and Ocln genes was co-transfected into mammalian human embryonic kidney (HEK) 293 cells and coimmunoprecipitation assay showed that DSP binds to Ocln, and binding of DSP to Ocln was narrow to DSPf5 and OclnL2 (Fig. 1J), whereas the DSP-Ocln interaction could be blocked by the DSP and Ocln antibodies, but not IgG as control (Supplementary Fig. 1). Using the gene runner computer software analysis of amino acids of the OclnL2 and DSPf5 reveals that these regions across many species are highly homologous (Supplementary Fig. 2A,B). This result indicates that DSP acts as a ligand and binds to its cell surface receptor, Ocln.
DSP interacts with occludin. Recombinant DSP 1–458, 1–213, 203–458, 1–89, 72–191, 182–295, 263–371, 363–458 were expressed in Escherichia coli BL21 and purified according to the manufacturer's instruction as described by “Materials and methods”. The purified DSP fusion proteins were confirmed by Coomassie blue staining (A,D) and Western blot assays using either an anti-GST or anti-DSP antibody (B). Interaction between DSP polypeptides and Ocln by GST pull down was detected by Western blot using anti-DSP and anti-Ocln antibodies (C,E,F). Arrow shows Ocln band (C). Mixture of different fragments of DSP and Ocln fusion proteins was pulled down by Ocln antibody and interaction of DSP with Ocln was detected using DSP antibody (E) and vice versa (F). The GST-DSP fusion protein constructs and the localization of the DSP protein stretch that contains Ocln-binding domain are illustrated (G). The DSP number starts at the translational start site of DSPP (Met) as No. 1. (H) Schematic representation of mouse Ocln protein structure from the initiation of the translation start site 1 to the end 522. OclnL1 and OclnL2 indicate the extracellular loop 1 (aa 85–183) and loop 2 (aa 194–241). (I) Either recombinant OclnL2 protein or BSA as control was coated in 96-well plates. Serial diluted biotinylated DSPf5 was added to the wells and incubated with the unlabeled recombinant OclnL2 protein or BSA, respectively. Bound DSPf5 was detected using AP-conjugated streptavidin and 1 mg/ml PNPP as substrate at 405 nm using a microplate reader. Binding affinity of different concentrations of DSPf5 to its substrate at the given time periods were calculated. Using the same method, binding of the biotinylated OclnL2 to the unlabeled DSPf5 was examined. Data point represent the mean ± S.D. (n = 3). (J) For in vivo studies, different fragments of DSP and Ocln cDNAs were subcloned into a CMV mammalian expression plasmid tagged with Flag or Myc peptides, respectively. Myc-Ocln and Flag-DSP as well as Myc-DSP and Flag-Ocln expression vectors were transfected into HEK-293 cells, respectively. After 48 h transfection, the proteins were harvested and protein-protein interactions were immunoprecipitated using Myc antibody. Protein-protein interaction was detected by Western blotting using anti-Myc, anti-DSP or anti-Ocln antibody.
Expression of DSP and Ocln during mouse tooth formation
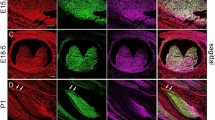
To determine whether the two proteins are coexpressed in tooth tissues, their distribution profiles during developing mouse teeth were examined using double labeling immunohistochemistry. At embryonic day (E) 13.5, DSP and Ocln expressions were not detected in craniofacial tissues (Fig. 2b,c). At postnatal day 1 (PN1), DSP was expressed in odontoblasts and weakly in ameloblasts (Fig. 2g). At PN5, DSP expression was strongly observed in odontoblasts and dentin and moderately in ameloblasts (Fig. 2l). Similar to PN5, DSP distribution exhibited the same expression pattern at PN15 in mouse teeth (Fig. 2q). Ocln signal coincided with DSP expression in odontoblasts (Fig. 2h,m,r), but Ocln expression pattern was also seen in stratum intermedium (SI). Although Ocln is an integral membrane protein, the NH2-and COOH domains of Ocln are localized in the cytoplasm22. Therefore, Ocln expression was observed in the cytoplasm besides the cell membrane. No signal was detected in the negative control tissue sections, where normal IgG was used instead of the primary antibodies of DSP and Ocln (Supplementary Fig. 3b,c).
Expression of dentin sialoprotein and occludin in developing mouse teeth. At embryonic day (E) 13.5, DSP and Ocln expressions were not detected in tooth tissues (b,c) using double fluorescent immunohistochemistry, but at postnatal day (PN) 1, DSP expression (red) was observed in odontoblasts, ameloblasts and dental pulp cells (g), Ocln expression (green) was detected in these areas overlapped with DSP (h). At PN5 and PN15, expression of DSP and Ocln was apparently seen in odontoblasts and ameloblasts (l,m,q,r). However, Ocln expression was relatively wider than that of DSP. (a,f,k and p) show bright images. The cells were stained with Hoechst for nuclei (d,i,n,s). Images were merged (e,j,o,t). (a–t) show higher magnifications from the yellow boxes.
DSP induces phosphorylation of Ocln and FAK and two protein expressions in odontoblasts
To elucidate whether DSP regulates modification of Ocln and its partner activity, mouse dental papilla mesenchymal cells were treated with DSPf5 protein. We found that DSPf5 phosphorylates Ocln at Ser490 at a dose-dependent manner (Fig. 3A,B). As several genes are Ocln partners29, we further investigated whether DSPf5 also regulates Ocln partner activity. Interestingly, we found that DSPf5 also induces FAK phosphorylation at Ser722 and Tyr576, but no effect on FAK at Tyr397 (data not shown) and Akt at Ser473 (Fig. 3A,B). Maximal induction of protein phosphorylation of Ocln-Ser490 and FAK-Ser722, FAK-Tyr675 by DSPf5 was at 2 h, 1 h and 1 h, respectively (Fig. 3C,D).
Effect of DSPf5 on occludin and FAK phosphorylation. (A) The mouse dental papilla mesenchymal cells were treated with or wither DSPf5 of 2 μg/ml (lane 2), 4 μg/ml (lane 3), 8 μg/ml (lane 4), 16 μg/ml (lane 5) and 24 μg/ml (lane 6) for 1 h at 37 °C. After cell harvest, protein expression was detected by Western blot assay using antibodies specific to p-Ocln-Ser490, Ocln, p-FAK-Ser722, p-FAK-Tyr576, FAK, p-AKT-Ser473, AKT and β-actin. (B) Protein expression levels were quantitated using imageJ software. Expression of p-Ocln-Ser490, p-FAK-Ser722, p-FAK-Tyr576 and p-AKT-Ser473 was normalized to Ocln, FAK and AKT, respectively. Protein on the control was considered as a one-fold increase. The expression level of proteins treated with DSPf5 was divided by protein level on the control group. The results showed the mean ± S.D. (n = 3). *p < 0.05; **p < 0.01. (C) The cells were treated with DSPf5 (8 μg/ml) in DMEM medium for 0. 15, 30, 60, 120 and 240 min. Protein expression was detected by Western blot analysis using antibodies specific to p-Ocln-Ser490, Ocln, p-FAK-Ser722, p-FAK-Tyr576, FAK, and β-actin, respectively. (D). Protein expression levels were quantitated using image J software. Expression of p-Ocln-Ser490, p-FAK-Ser722 and p-FAK-Tyr576 was normalized to Ocln and FAK, respectively. Protein treated with DSPf5 at 0 min as control group was considered as a one-fold increase. The expression level of proteins treated with DSPf5 at different time periods was divided by protein level on the control group.
To determine relationship between Ocln and FAK within odontoblasts during odontogenesis, expression of Ocln and FAK in odontoblasts was performed using double labeling fluorescent immunohistochemistry. Our study showed that expression of Ocln and FAK was observed in odontoblasts at PN1, 5, and 10 (Fig. 4A). No signals were detected in the control tissue sections when the antibodies of Ocln and FAK were replaced by normal IgG as negative control (Supplementary Fig. 4b,c). Also, expression of Ocln and FAK proteins was seen in mouse dental papilla mesenchymal cells (Fig. 4B) and HEK 293 cells (Supplementary Fig. 4g,h), but no immunostaining was seen in the control groups (Supplementary Fig. 4l,m,q,r). To further elucidate if Ocln interacts with FAK, mammalian expression vectors of Ocln and FAK genes were transfected into mammalian cells. Coimmunoprecipitation assay demonstrated that Ocln physically interacts with FAK in vivo (Fig. 4C).
Expression of occludin and FAK in odontoblasts during tooth development. (A) Fluorescent immunohistochemistry showed that at PN1, 5, 10 during mouse tooth development, Ocln (red) and FAK (green) were co-expressed in odontoblasts by the double immunofluorescent histochemistry using antibodies specific to Ocln and FAK (b,c,g,h,l,m). (a, f and k) show bright images. The cells were stained with Hoechst for nuclei (d,i,n). Images were merged (e,j,o). Am, Ameloblasts; Od, Odontoblasts. (B) Ocln (red) and FAK (green) were expressed in iMDP-3 cells using double immunofluorescent assay (b,c). a shows bright image. Hoechst was used for nuclear staining (d). Image was merged (e). (C) Ocln interacts with FAK in vivo. Myc-Ocln and GFP-FAK mammalian expression vectors were cotransfected into HEK-293 cells. After 48 h transfection, the cells were harvested. Ocln and FAK were immune-precipitated using Myc or GFP antibody. Interaction of Ocln with FAK was detected by Western blot assay using Ocln and FAK antibodies.
Effect of DSPf5 on phosphorylation of Ocln and FAK is disrupted by DSP and Ocln antibodies
To further assess phosphorylation of Ocln and FAK mediated by DSP-Ocln signaling, we sought to block DSP signal pathway using DSP and Ocln antibodies. Mouse dental papilla mesenchymal cells were treated by DSPf5 with or without the DSP or Ocln antibody and the cells were harvested for immunoblot analysis. It was noted that the DSP and Ocln antibodies could block phosphorylation of Ocln at Ser490 and FAK at Ser722 and Tyr576 compared to the control group (Fig. 5A-D), but IgG as control had no effect on block of the two protein phosphorylations induced by DSPf5. Also, disruption of the two protein phosphorylation by the DSP and Ocln antibodies was dose-dependent. With increase of the DSP and Ocln antibodies, phosphorylation of p-Ocln at Ser490, p-FAK at Ser722 and p-FAK at Tyr576 was dramatically decreased. Immunohistochemistry further indicated that phosphorylated levels of the two proteins were attenuated by the DSP and Ocln antibodies (Fig. 6A–C; Supplementary Fig. 5), indicating that DSP regulates phosphorylation of Ocln and FAK proteins via DSP-Ocln signaling.
Effect of DSPf5 on occludin and FAK phosphorylation is blocked by DSP and occludin antibodies. (A,C) The mouse dental papilla mesenchymal cells were treated with or without DSPf5, or DSPf5 plus DSP antibody or DSPf5 plus Ocln antibody or DSPf5 with IgG as control for 1 h, respectively. Protein expression was detected by Western blotting using antibodies described above. Lanes, 1 and 6 as control without the DSPf5 induction; lanes 2 and 7 (8 μg/ml of DSPf5); lane 3 (8 μg/ml of DSPf5 plus 8 μg/ml of DSP antibody); lane 4 (8 μg/ml of DSPf5 plus 16 μg/ml of DSP antibody); lane 8 (8 μg/ml of the DSPf5 plus 8 μg/ml of Ocln antibody); lane 9 (8 μg/ml of DSPf5 plus 16 μg/ml of Ocln antibody); lanes 5 and 10 (8 μg/ml of DSPf5 plus 16 μg/ml of IgG). (B,D) Protein expression level was measured using imageJ software. Expression of p-Ocln-Ser490, p-FAK-Ser722 and p-FAK-Tyr576 was normalized to Ocln and FAK, respectively. Protein without treatment was considered as one-fold increase. The expression protein level of the treated groups was divided by protein level on the control group. The results are shown as the mean ± S.D. (n = 3). *p < 0.05; **p < 0.01.
DSP and occludin antibodies block phosphorylation of occludin and FAK proteins mediated by DSPf5 in mouse dental papilla mesenchymal cells. (A–C) The cells were treated with or without DSPf5 or DSPf5 (16 μg/ml) plus the DSP (8 μg/ml) or Ocln (8 μg/ml) antibody for 1 h at 37 °C. The cells were fixed and immune-stained using p-Ocln-Ser490 (A), p-FAK-Ser722 (B) and pFAK-Tyr576 (C) antibodies, respectively. Data showed that effect of DSPf5 on Ocln and FAK phosphorylation was blocked by the DSP or Ocln antibody (e–h). (a–d) are bright images. The cells were stained with Hoechst for nuclei (i–l). Images were merged (m–p).
DSP induces dental cell differentiation and mineralization
As DSP is abundant in dentin ECM during odontoblast differentiation, we further addressed the possibility that Ocln-FAK signaling could be involved in controlling functional activities of dental cell differentiation and mineralization. To directly assess DSP effect on dental cell differentiation, we firstly examined differentiation of human dental pulp stem cells and mouse dental papilla mesenchymal cells in cultured systems. ALP assay revealed that DSPf5 has high effect on human dental pulp stem cell and mouse dental papilla mesenchymal cell differentiation compared to the control group (Fig. 7A–D). Alizarin red S assay also demonstrated that DSPf5 stimulates more calcium deposition of the two dental cells after 14 days of culture than that of the control groups (Fig. 7E–H). The cell differentiation and mineralization induced by DSPf5 protein was attenuated by the DSP and Ocln antibodies, whereas IgG as control had no effect on the cell differentiation and mineralization stimulated by DSPf5 (Supplementary Fig. 6). Furthermore, gain- and loss-Ocln gene in the mouse dental papilla mesenchymal cells up- and down-regulated the cell differentiation and mineralization (Supplementary Fig. 7). The data indicate that DSP promotes the dental cell differentiation and mineralization partially through the Ocln signaling.
Effect of DSP domain on dental cell differentiation and mineralization. Mouse dental papilla mesenchymal (iMDP-3) (A) and human dental pulp stem (hDPSC) cells (B) were treated with or without 10 μg/ml of DSPf5 in calcifying medium for 7 days. ALP activity was analyzed using in situ ALP staining (A, B). (C,D) Quantitative ALP activity of the cell lysates was assayed using ρ-nitrophenyl phosphate as a substrate. Protein concentration was determined using the BCA protein assay reagent as described in “Materials and methods”. There were significantly different between the DSPf5 treated groups and DSPf5 untreated groups. For cell biomineralization, iMDP-3 (E) and hDPSC cells (F) were maintained in the same condition for 14 days. The cells were fixed and stained for Alizarin red S dye. (G,H) The amount of calcium deposition was quantified by destaining with 10% cetylpyridinium chloride in 10 mM sodium phosphate at room temperature. Mineralization deposits were determined and data represent mean ± S.D. (n = 3). Con, control. *p < 0.05; **p < 0.01.
To further assess whether this DSP peptide modulate endogenous dental mesenchymal cell differentiation and biomineralization in an in vivo context, we mixed the peptide with Affi-Gel Blue Gel. Data showed that DSPf5 highly binds to the Affi-Gel Blue Beads, forming a DSP-bead compound and this DSPf5 peptide is also easily released from the compound (Fig. 8A). Then, the compound was implanted into mouse dental pulp chambers. Tissue morphology after 1-, 3-, and 5-week surgery was observe by histological chemistry, and H&E stained tooth tissue sections revealed that there were not significant differences of tooth morphology between the control and DSPf5 treated groups after 1 week surgery (Supplementary Fig. 8a–f). However, after 3 week treatment of DSPf5, DSPf5 was able to induce the dental pulp mesenchymal cell proliferation and differentiation. Dental pulp mesenchymal cells in the DSP-treated group secrete dental ECM at the top “artificial hole” between the resin and dental pulp chamber (Fig. 8Ba-g). More interestingly, in the DSP-treated group, there were many blood vessels and less inflammatory cells around the agarose beads and dental pulp cells. Also, the agarose beads were resorbed as well as dental pulp cells and blood vessels were invaded into the agarose beads. Similar to the 3-week DSP treatment group, in the 5-week DSP induced groups, ECM covered major space at the wound area and less inflammatory cells were seen in the dental pulp cavity. Many blood vessels were apparent around the agarose beads (Fig. 8Ca–h). However, formation of reparative dentin was not seen in the DSP treated groups, the mechanism of the DSP domain in the reparative dentin formation needs to be further investigated in the future. These findings indicate that the DSP domain induces differentiation of dental pulp mesenchymal cells into odontoblast-like cells and differentiating cells are capable of synthesizing and secreting ECM.
Effect of DSP domain on dental cell differentiation in vivo. (A) Lane 1, DSPf5 protein only; lane 2, DSPf5 was mixed with Affi-Gel Blue Gel Beads at room temperature for overnight. The mixture was centrifuged and supernatant was loaded onto a 7% SDS-PAGE gel; lane 3, the pellet was added to 1 x SDS-loading buffer and heated. The released DSPf5 from the compound was loaded onto a 7% SDS-PAGE gel. (B) The complex of DSPf5 coated to agarose beads was implanted into mouse dental pulp chambers. Compared to the control group after 3 week operation (Ba–c), in the DSPf5-treated group, DSPf5 was able to induce the dental pulp cell proliferation and differentiation. Dental pulp cells in the DSPf5-treated group secrete ECM (arrow) at the top “artificial hole” between resin and dental pulp. Blood vessels (BV) proliferate and migrate near agarose beads (AB). Arrowheads show that dental pulp cells differentiate and secrete ECM. b, c and e–g are enlarged from the boxes in the b’, c’ and e’–g’. In DSPf5-treated group after 5 week operation (C), ECM secreted by dental pulp cells forms a layer covering “the artificial hole” between resin and dental pup cells (Ce, f). Dental pulp cells surround the agarose beads (AB) and AB was resorbed and dental pulp cells and blood vessels invade into AB (Cg, h). A number of inflammatory cells were decreased compared to the control group. b–d and f–h are enlarged from the boxes in the b’–d’ and f’–h”. B, alveolar bone; BV, blood vessels; DP, dental pulp cells; ECM, extracellular matrix; IC, inflammatory cells; R, resin; T, teeth.
Discussion
DSP is highly expressed in dentin ECM during odontoblast differentiation and dentin formation. Its mutations are associated with dentin hereditary diseases19,20,21, 44. Previous studies showed that DSP and its fragments derived from DSP regulate intracellular signal transductions9, 16, 26. Unlike other SILING members, DSP domain does not contain RGD8, 45. Therefore, mechanisms of DSP in regulation of intracellular signaling during dentinogenesis have not well been understood. Recently, our and other laboratories found that DSP is processed by MMP20 and MMP9 into the given fragments11, 15 and the middle domain of DSPaa 183–219 binds to integrin β6, forming a complex. This complex phosphorylated transcription factors, Smad1/5/8, through p38 and Erk1/2 protein kinases and up-regulated expression of DSPP and DMP1 genes as well as induced dental mesenchymal cell attachment, differentiation and mineralization9. In this study, we used DSP as bait to screen protein library from mouse odontoblast-like cells and identified that several cell membrane proteins were bound by DSP including Ocln (Table 1). Previous studies showed that Ocln interacts with several cellular molecules and activates intracellular signal transductions22, 29,30,31. Ploss et al. identified that Hepatitis C virus (HCV) enter human hepatic cells through HCV glycoproteins (HCVpp) and OCLN is an essential entry factor. They furthermore found that the specific determinants of OCLN’s HCV entry factor functions are entirely contained within the OCLN extracellular loop 231. As DSP is able to bind to Ocln in vitro and in vivo, we further studied which domain of DSP interacts with Ocln and found that the COOH-DSP domainaa 363–458 is a ligand and binds to the extracellular loop 2 (OclnL2) of Ocln. This DSP domain regulates intracellular signaling through a direct matrix-cell interaction. Immunohistochemistry showed that expression of DSP and Ocln is stage-specific in odontoblasts during dentinogenesis.
Ocln is an integral membrane protein expressed in epithelial and other cells and was originally predicted to confer barrier properties to the TJs27,28,29. During dentinogenesis, adjacent odontoblasts are tightly attached together and freeze-fracture studies showed that TJs between odontoblasts appear as short rows of fused particles during early mantle dentin mineralization and become more complex, eventually forming networks of fused particles in advanced mineralization46,47,48,49. The transport of calcium ions through odontoblasts is mediated by specific cell organelles and cell membrane domains47, 48, 50,51,52,53. TJs between odontoblasts are mainly composed of transmembrane proteins including claudin and Ocln as well as cytoplasmic proteins [such as zona occludens-1 (ZO-1), ZO-2 and ZO-3, FAK]48, 49, 54. Expression of the TJ-associated proteins was clearly detected in odontoblasts, however, temporal and spatial distributions of these proteins such as Ocln in differentiating odontoblasts are different48, 49, 55,56,57,58. The cause of the differences is still uncertain, and it may be due to the animals used and the methods for specimen preparation as well as antibody concentrations in each study59. Lee et al. observed that Ocln and ZO-1 proteins are strongly expressed in odontoblasts at PNs from 7 to 18 examined in the wild type mouse teeth whereas expression of Ocln and ZO-1 is hardly detected in odontoblasts of Nfic-deficient mice at the same ages. Nfic-deficient teeth failed to differentiate into normal odontoblasts and these aberrant odontoblasts were round and dissociated and lost their polarity. Electron microscopy of aberrant odontoblasts exhibited no intercellular junctional complex between them58. Lungrw et al. described that MRPC-11 cells derived from rat dental pulp cell line exhibit an odontoblast-like phenotype and express DSPP and Ocln. TJ complex was seen in the intercellular spaces of these cells and the transcellular Ca2+ flux was inhibited by nifedipine, giving evidence for an active intracellular Ca2+ transport through voltage-gated channels60. In this study, we observed that expression of DSPP/DSP and Ocln was not detectable in tooth tissues at embryonic days 13.5 (Fig. 2Ab,c). With odontoblast cytodifferentiation, both of these proteins were expressed in odontoblasts at PNs from 1 to 15 examined (Fig. 2Ag,h,l,m,q,r). Our results are in agreement with previous studies by other groups5, 55,56,57,58,59,60,61.
Ocln mutations in humans cause band-like brain calcification, polymicrogyria and advanced chronic renal disease15, 33,34,35. Ocln deficient mice developed deafness with dislocalization of tricellulin in cochlea36. Tricellulin is a recently identified constituent of TJs, and is the first marker of the tricellular tight junction (tTJ)62. Also, Ocln knock-out mice showed calcification in the brain, testicular atrophy, loss of cytoplasmic granules in striated duct cells of the salivary gland, and thinning of the compact bone, but in Ocln knockout-mice, morphology of the TJs does not appeared to be affected and barrier function of epithelium is normal37 and cells originating from Ocln-deficient embryonic stem cells have well-developed networks of tight-junction strands38, suggesting that the functions of Ocln are more complicated than previously supposed. These contradictory data led us to ask if Ocln contributes to a regulatory function in intracellular signal transductions. Our current study demonstrated that a novel role of Ocln acts as a receptor interacting with DSP for modulating intracellular functions via DSP-Ocln-FAK axis. It was found that several ECMs and molecules interacting with either extracellular loop 1 or 2 of Ocln regulate intracellular signaling through phosphorylation/dephosphorylation of Ocln28,29,30,31, 63. In the present study, we found that DSPaa 363–458 binds to the extracellular loop 2aa 194–241 of Ocln. Both of the DSPaa 363–458 and OclnL2aa 194–241 are high homologous across species lines. In human, the mutation of DSP at codon 45 (c.133C > T, p.Q45X) introduces a premature terminal signal and would results in a truncated protein without the COOH-terminal DSP domain21, 25, 64, 65. As the mouse COOH-terminal DSP domainaa 363–458 is highly homologous to the human COOH-terminal DSP domainaa 374–46966 and this domain regulates dental mesenchymal cell lineages and mineralization through Ocln-FAK signal. It is assured that the lack of the domain of the human DSP in these cases (p.Q45X) may be relevant to DGI.
Evidence for Ocln phosphorylation has previously been described28,29,30. However, the mechanisms by which these modifications through DSP influence intracellular signaling during dentinogenesis have not been elucidated. We demonstrated that the DSP peptide phosphorylates Ocln at Ser490 in mouse dental papilla mesenchymal cells and the Ocln phosphorylation at Ser490 induced by DSPf5 was blocked by DSP and Ocln antibodies, implying Ocln phosphorylation at Ser490 through DSP signaling. Previously, it was found that vessel endothelial growth factor (VEGF) induces Ocln phosphorylation at Ser490 through activating protein kinase C (PKC) β and regulation of vascular permeability63. We observed that besides induction of Ocln phosphorylation, DSP also activated FAK phosphorylation at Ser722 and Tyr576 in mouse dental papilla mesenchymal cells. Maximal induction of Ocln and FAK phosphorylation was at 1- and 2-h of DSPf5 treatment. FAK is ubiquitously expressed in a variety of cells. Previous study showed that FAK is structurally associated with Ocln, but not other tight junction proteins, JAM-A and claudin-11 in Sertoli cells. FAK knock-down resulted in a significant loss of Ocln-ZO-1 interaction and interruption of the TJs67. Our studies demonstrated that similar to coexpression of DSP and Ocln, expression of both Ocln and FAK is stage-specific in odontoblasts during tooth development and in mouse dental papilla mesenchymal cells as well as Ocln physically interacts with FAK (Fig. 4). Expression of Ocln and FAK in odontoblastic cells during tooth formation was also observed by other groups24, 58, 60. Thus, coexpression of the three of DSP, Ocln and FAK exists in odontoblasts during tooth development. FAK bridges the cytoskeleton on the inside of cells with components of the ECM on the outside of cells via the cell surface receptors such as integrin and others68, 69. FAK protein contains many phosphorylated sites and phosphorylation events occurring within FAK influence numerous processes including mitogenic signaling, cell migration, proliferation and differentiation. Its activity is modulated by phosphorylation, either in a positive or negative fashion70, 71. FAK phosphorylation could be induced by adhesion of cell surface receptors such as integrin to ECM and by a variety of other extracellular factors72. Our study demonstrated that FAK phosphorylation at Ser722 and Tyr576 was blocked by either DSP or Ocln antibody. These findings suggest that FAK is a substrate of Ocln. However, the mechanism how Ocln phosphorylates FAK at Ser722 and Tyr576 remains unknown and whether the DSP affects binding affinity and structure of Ocln and FAK needs to be further investigated. In fact, recent studies have found that there are multiple potential Tyr, Ser and Thr phosphorylation sites in FAK mapped by mass spectrometry73.
In this study, we found that DSP domainaa 363–458 interacts with the extracellular loop 2 of Oclnaa 194–241. The DSP domain induced phosphorylation of Ocln at Ser490 and FAK at Ser722 and Tyr576. The phosphorylation of Ocln and FAK proteins could be blocked by the anti-DSP and anti-Ocln antibodies, respectively. Immunohistochemistry analysis showed that co-expression of DSP, Ocln and FAK proteins are present in mouse odontoblasts and dental mesenchymal cells. Furthermore, in vivo study revealed that Ocln binds to FAK, indicating that the DSP domain regulates the intracellular activities through the Ocln-FAK signaling pathway. For the functional study, we observed that the DSP peptideaa 363–458 is sufficient to induce differentiation and mineralization of human dental pulp stem cells and mouse dental papilla mesenchymal cells in the cultured system. Effect of DSP on dental papilla mesenchymal cell differentiation and mineralization could be blocked by the DSP and Ocln antibodies and gain- and loss-Ocln gene in these cells was able to up- and down-regulate the cell differentiation and mineralization. More importantly, when this peptide was implanted into mouse dental pulp chambers, this DSP peptide was able to induce endogenous dental pulp cell proliferation, differentiation. The differentiating dental pulp cells were capable of synthesizing and secreting dental ECM and migrating to the region between resin and dental pulp chamber as well as new synthesized dentin ECM covered the wound area. Also, in DSP-treated groups, more blood vessels were seen surrounding dental pulp cells and agarose beads. Less inflammatory cells were observed in dental pulp chambers compared to the control groups. Although the formation of dentin extracellular matrix between the wound area and dental pulp chamber in the DSP treated groups in 3- and 5-week surgery was seen, the formation of the reparative dentin in both the two groups was not observed. The phenomenon remains to be further studied. In the present study, these findings suggest that this DSP peptide could be a potentially interesting therapeutic molecule by locally regenerating dentin-pulp complex. However, the detail mechanisms by which DSP domain mediates dental cell proliferation, differentiation, migration and blood vessel regeneration through Ocln-FAK signaling transduction needs to be further investigated in the future. Figure 9 depicts a model by which DSP domainaa 363–458 regulates dental cell differentiation and mineralization via DSP-Ocln-FAK signaling based on this report.
DSP domain regulates dental mesenchymal cell differentiation through occludin-FAK signaling. The hypothetical model depicts that the DSPaa 363–458 acts as a ligand and interacts with the extracellular loop 2 of Oclnaa 194–241, activating Ocln phosphorylation at Ser490. Furthermore, the DSP-Ocln complex activates FAK phosphorylation at Ser722 and Tyr576 and then induces dental mesenchymal cell differentiation and mineralization.
Materials and Methods
Antibodies and Reagents
A rabbit polyclonal anti-mouse DSP antibody recognizes residues between ILe18 and Lys371, (M-300), #sc-33587; goat polyclonal anti-mouse DSP (M-20), #sc-18328; rabbit polyclonal anti-mouse p-FAK (Tyr-397), #sc-11765-R; rabbit polyclonal anti-mouse p-FAK (Tyr-576), #sc-16563-R; mouse monoclonal anti-mouse p-FAK (Ser-722), #sc-374668; rabbit polyclonal anti-mouse FAK, #sc-557; rabbit polyclonal anti-Akt, #sc-5298; rabbit polyclonal anti-mouse p-Akt1/2/3 (Ser-473), #sc-7985-R; goat polyclonal actin, #sc-1616 antibodies, mouse Ocln shRNA plasmid #sc-36118 were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). A polyclonal anti-DSP-COOH antibody was produced in rabbit using the oligopeptide with the sequence of KRNSPKQGESDKPQGTAE (mouse DSP residues 401–418; Alpha Diagnostic International, San Antonio, TX, USA, #2603). Monoclonal anti-Myc-, #13–2500; anti-GFP, #33-2600; and anti-Flag, #MA1-91878 antibodies were purchased from Thermo Fischer Scientific (Waltham, MA, USA). A monoclonal occludin antibody was obtained from Life Technologies (Carlsbad, CA, USA, #33-150). A rabbit antibody against occludin phosphorylated at Ser-490 was kindly provided by Dr. Antonetti (Department of Cellular and Molecular Physiology and Ophthalmology, Penn State College of Medicine, Pennsylvania, USA). Normal mouse immunoglobulin G (IgG) (Vector Laboratories, Burlingame, CA, USA, #I-2000) was employed as a negative control.
Animals and tissue preparation
All experimental procedures involving the use of animals were approved by the University of Texas Health Science Center at San Antonio (UTHSCSA, approval number: IACUC-50012X), TX, USA. ICR mice were purchased from Harlan-Laboratory Animals Inc. (Indianapolis, IN. USA). The tissue sections were prepared as previously described in detail74. Mice with litters of embryonic day (E) 13.5 and postnatal days (PN) 1, 5, 10 and 15 were sacrificed. Mouse tissues were dissected and fixed in 4% paraformaldehyde overnight. After demineralization in 10% EDTA, samples were dehydrated in increasing concentrations of ethanol, embedded in paraffin. Serial sections (4 μM) were prepared for immunohistochemistry analysis. All experimental methods were conducted in accordance with relevant guidelines.
Cell culture
Mouse dental papilla mesenchymal cells75 and human dental pulp stem cells76 were grown in alpha minimal essential medium (a-MEM) (Life Technologies, USA) with 10% fetal calf serum (FCS, Atlanta Biologicals Inc., Norcross, GA, USA), 50 µg/ml ascorbic acid, 10 mM sodium β-glycerophosphate, 100 units/ml penicillin/streptomycin (Sigma–Aldrich, St. Louis, MO, USA) at 37 °C under 5% CO2. Human embryonic kidney (HEK-293) cells were purchased from American Tissue Collection Center (ATCC, Manassas, VA, USA; #CRL-1573) and were maintained in Dulbecco’s modified eagle medium (DMEM) with 10% FCS, 100 units/ml penicillin/streptomycin at 37 °C under 5% CO2.
Generation of recombinant dentin sialoprotein and occludin
The expression and purification of a recombinant DSP were performed according to previously described protocols16. Full-length and different fragments of mouse DSP cDNA were amplified by PCR using a full-length mouse DSPP cDNA as a temple with primers shown in Supplementary Table 2 adding EcoRI sites at both ends for directional ligation into the expression vector pGEX-6P3 with EcoRI sites (Amersham Biosciences, Piscataway, NJ, USA) and named DSP, DSP-N, DSP-C, DSPf1, DSPf2, DSPf3, DSPf4 and DSPf5 (Fig. 1G). Mouse Ocln plasmid (pBluescript SK-Ocln) was kindly provided by Dr. Mikio Furuse (Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Hyogo, Japan). Full-length and different fragments of Ocln cDNA including the whole length and extracellular loops 1 (OclnL1) and 2 (OclnL2) of Ocln gene were amplified by PCR using a full-length mouse Ocln cDNA as a template. Primers used for generation of Ocln DNA constructs were shown in Supplementary Table 2. PCR products were subcloned into pGEX-6P3 vector, respectively (Amersham Biosciences, USA). After confirming the right sequence, the resulting plasmid was transformed into Escherichia coli BL21. The protein expression and purification were performed according to the manufacturer’s instruction (Amersham Biosciences, USA). Briefly, the fusion protein was induced by the addition of 1 mM of isopropyl b-D-thiogalacto pyranoside (IPTG) at 37 °C for 5 h. The recombinant DSP and Ocln proteins were then purified using a GSTrap 4B protein purification system (Amersham Biosciences, USA, #28-4017-45). The purified recombinant proteins were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), followed by Coomassie brilliant blue staining and Western blotting analysis using GST antibody (1:2,000, Amersham Biosciences, USA, #27-45-77-01). For in vivo study, DSP and Ocln genes were subcloned into CMV expression vector tagged with either Myc or Flag (Sigma-Aldrich, USA, #pp2393, #pp2395). Mammalian mouse FAK with GFP expression plasmid was obtained from Addgene (Cambridge, MA, USA, #50515).
Glutathione fusion protein (GST) pull down assay
GST pull down assay was according to the manufacture’s instruction (Amersham Biosciences, USA). In brief, GST-DSP fusion protein was incubated with 1 ml of cell lysis from mouse odontoblast-like cells (MO-6G3)6 in the cold lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, Triton X100 (Sigma-Aldrich, USA) overnight at 4 °C. After the reaction, the 50 μl of a 50% slurry of glutathione agarose beads (Amersham Biosciences, USA) were added for further incubation for 2 h at 4 °C. The samples were centrifuged at 12,000 g for 2 min at 4 °C and the supernatant was removed. After extensive washes containing 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.025% sodium azide, the DSP binding proteins were eluted by 20 mM reduced glutathione in 50 mM Tris-Cl, pH 8.0. The eluted samples (10 μg) were mixed with 2x SDS-PAGE gel loading buffer after boiling and run onto 7% SDS-PAGE gels and were transferred to Trans-Blot membranes (Bio-Rad Laboratories Inc., Hercules, CA, USA). The membrane was blocked with 5% non-fat milk in TBST buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1% Tween-20) for 60 min at room temperature. After blocking, the membranes were incubated overnight at 4 °C with the respective primary antibodies (1:500-1,000 dilutions) shown in Table 1 and Supplementary Table 1. The membranes were washed with 1x TBST and incubated with diluted horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000, Pierce, Rockford, IL, USA) for 1 h at room temperature. After three washes, the membranes were detected using an enhanced chemiluminescence (ECL) kit (Millipore, Bedford, MA, USA).
Co-immunoprecipitation
Co-immunoprecipitation assay was performed according to the manufacture’s instruction (Active Motif, Carlsbad, CA, USA, #54002). Briefly, HEK 293 cells from a 100-mm Petri dish (70–90% subconfluent) were transfected with 20 μg of Flag and c-Myc plasmids containing DSP and Ocln genes using the Lipofectamine 2000 (Life Technology, USA). After 48 h transfection, the cells were lysed in 3 mL of lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and proteinase inhibitor cocktail (Sigma-Aldrich, USA) and mixed with a vortex mixer for 1 h at 4 °C. Insoluble materials were removed by centrifugation at 12,000 g for 10 min at 4 °C. Lysates containing 5 mg of proteins in 1 mL were precleared by incubating with 40 μl of anti-FLAG or anti-Myc Affinity Gel (Sigma-Aldrich, USA, #A4596 and #IP0020) at 4 °C on a rotator overnight. Immunoprecipitates were washed three times with 1 x TBST and heated for 5 min at 95 °C in 20 µl 2x SDS-PAGE sample buffer (62.5 mM Tris HCL, pH 6.8. with 2%SDS, 10% glycerol, 0.002% bromophenol blue). The bound materials were resolved on 7% SDS-PAGE gels for analysis by Western blotting with anti-Ocln (1:1,000, Life Technologies, USA), or anti-Myc (1:1,500; Thermo Fischer Scientific, USA) or anti-DSP (1:1,000, Santa Cruz Biotechnology Inc., USA) antibody. In vitro immunoprecipitation of DSP and Ocln fusion proteins was performed according to the manufacture’s protocol (Amersham Biosciences, USA). In brief, 10 μg of GST-DSP and - Ocln fusion proteins were pre-cleared with 10% protein G-strepharose in binding buffer. The samples were then incubated with 10 μg of either primary DSP (Santa Cruz Biotechnology Inc., USA) or Ocln (Life Technologies, USA) antibody, followed by immunoprecipitation with 20 μl protein G-strepharose (Amersham Biosciences, USA). Immunoprecipitations were washed extensively with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) and boiled for 5 min. The supernatants were separated by 7% SDS-PAGE gels, transferred to nitrocellulose membranes, immunoblotted with primary DSP (1:1,000, Santa Cruz Biotechnology Inc., USA) or Ocln (1:1,500, Life Technologies, USA) antibody at 4 °C overnight. The membranes were then washed with 1 × TBST for three times and incubated with diluted horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000, Pierce, USA) for 1 h at room temperature. After three washes, the membranes were detected using an enhanced chemiluminescence (ECL) kit (Millipore, USA).
Biotinylation of dentin sialoprotein and substrate binding assay
For probing protein–protein interactions in vitro, recombinant DSPaa 363–458 (DSPf5) and Ocln loop2aa 194–241 (OclnL2) proteins at concentrations of 300 mg/ml was dialyzed against 0.1 M NaHCO3 and then reacted with 100 mg/ml Sulfo-NHS (N-hydroxysuccinimido)-LC (long-chain)-biotin according to the manufacture’s instruction (Pierce, USA) for 20 min at room temperature, followed by 2 h at 4 °C, respectively. Free biotin was removed by dialysis against 50 mM Tris-HCl and 150 mM NaCl, pH 7.4. To characterize the relative DSPf5 interaction with OclnL2, substrate binding assay was performed as described earlier77. Briefly, 96-microwell plates were coated with 1 μg/per well of OclnL2 and bovine serum albumin (BSA) as control overnight at 4 °C and non-specific binding sites were blocked with 1% BSA (Sigma-Aldrich, USA) at room temperature for 1 h. After through rinses with 1x PBS, the biotinylated DSPf5 ranged from 0–18 fold molar excesses in PBS with 1% BSA was added into the plates and then incubated at room temperature for different time periods. Bound DSPf5 was reacted with AP-conjugated streptavidin diluted 1:10,000 in 1x PBS for different time points at room temperature using 1 mg/ml PNPP (ƿ-nitrophenyl phosphate disodium) as substrate (Pierce, USA) and quantified at 405 nm (Opsys MR, Dynex, Chantilly, VA, USA). BSA was used as a control group and the binding of DSPf5 to OclnL2 was expressed as relative binding activity compared to the control group. At the same way, binding of the biotinylated OclnL2 to the unlabeled DSPf5 was performed. All experiments were performed from three independent experiments in triplicate.
Protein sequences and data analyses
A database search was performed at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast) using the BLAST program. The DSPP and Ocln nucleotides and derived amino acids were aligned with those from different species using the Gene Runner software program (http://www.generunner). Accession numbers of mRNA and protein sequences of DSPP and Ocln were shown in Supplementary Fig. 2.
Alkaline phosphatase (ALP) and mineralization assays
For detection of ALP activity, cultures of the human dental pulp stem and mouse dental papilla mesenchymal cells at 6-well palates with 104 cells per well were treated with or without DSPf5 (10 μg) induction in calcifying medium (α-MEM supplemented with 3% FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml), 50 μg/ml ascorbic acid, 10 nM dexamethasone and 10 mM sodium β-glycerophosphate) at 37 °C for 7 days and fixed with 70% ethanol for 5 min and washed with 1 x PBS. In situ ALP staining was performed according to the manufacture’s instruction (Bio-Rad Laboratories Inc., USA). Quantitative ALP activity of the cell lysates was assayed using ρ-nitrophenyl phosphate as a substrate. Protein concentration was determined using the BCA protein assay reagent (Pierce, USA). The enzyme activity was expressed as nanomoles of ρ-nitrophenol produced per min per mg of protein. For mineralization assay, calcium deposition was determined according to the manufacture’s recommendation protocol (Sigma–Aldrich, USA). In brief, these cells were seeded into 6-well plates and treated with or without 10 μg of DSPf5 in calcifying medium at 37 °C for 14 days. The cells were fixed in 10% formaldehyde neutral buffer at room temperature for 15 min. The monolayers were then washed twice with excess ddH2O prior to addition of 1 mL of 40 mM alizarin red S dye (pH 4.1) per well (Sigma–Aldrich, USA). The plates were incubated at room temperature for 20 min with gentle shaking. After aspiration of the unincorporated dye, the wells were washed four times with ddH2O while shaking for 5 min. Stained monolayers were visualized by phase microscopy using a Nikon Eclipse TE2000S inverted microscope with a filter by means of a digital cooled camera connected to a PC computer and analyzed with NIS-Elements 3.2 software (Melville, NY, USA). The amount of calcium deposition was quantified by de-staining with 1 ml of 10% cetylpyridinium chloride (Sigma–Aldrich, USA) in 10 mM sodium phosphate at room temperature for 1 h. The dye was then removed and 200 μL aliquots were transferred to a 96-well plate prior to reading at 550 nm using a microplate reader (Opsys MR, USA). Experiments were performed in triplicate and repeated in three cultures (n = 3).
RNA preparation and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from the mouse dental papilla mesenchymal cells using RNA STAT-60 kit (Tel-Test, Inc. Friendswood, TX, USA, #cs-110), treated with DNase I (Promega, Madison, WI, USA, #M6101), and purified with the RNeasy Mini Kit (Qiagen Inc., Valencia, CA, USA, #74104). RNA concentration was determined at an optical density of OD260. The RNA was transcribed into cDNA by SuperScript II reverse transcriptase according to the manufacture’s instruction (Life Technologies, USA, #18-080-044). qPCR was performed in a 20 μl reaction containing 2 ρmol of specific primers as follows: DSPP forward, 5′-AACTCTGTGGCTGTGCCTCT-3′; reverse, 5′-TATTGACTCGGAGCCATTCC-3′; Ocln forward, 5′-CATAATGGGAGTGAACCCGAC-3′, reverse, 5′-TATAGCCTCCTGGGGATC-3′. The PCR reaction was first denatured at 95 °C for 5 min, and then carried out at 95 °C for 60 s, at 55 °C–60 °C for 60 s and at 72 °C for 60 s for 30 cycles and with a final 10 min extension at 72 °C in T100TM Thermal Cycler (Bio-Rad Laboratories Inc., USA, #1861096). Five μl of PCR products were analyzed by 1.5% agarose gels with ethidium bromide staining.
Immunohistochemistry
In order to examine the expression of DSP, Ocln and FAK during tooth development and dental papilla mesenchymal cells, the fluorescent immunohistochemistry was performed according to previous descriptions74. In brief, tissue sections were dewaxed in xylene for 15 min for 3 times, hydrated in gradually decreasing ethanol concentrations for 2 min each and immersed in ddH2O for 5 min for three times. Antigen retrieval was performed by treating the sections with 0.1% (w/v) trypsin (Sigma-Aldrich, USA) at room temperature for 15 min. The sections were successively pretreated with 0.3% hydrogen peroxide for 30 min, blocked with 10% normal donkey serum (Sigma–Aldrich, USA). For double labeling, two primary antibodies were incubated simultaneously overnight at 4 °C at the following dilutions: rabbit anti-FAK (1:100; Santa Cruz Biotechnology Inc., USA, #sc-557), goat anti-DSP (1:100; Santa Cruz Biotechnology Inc., USA, #sc-18328), mouse anti-Ocln (1:100; Life Technologies, USA, #33-150), which were recognized by donkey anti-rabbit, anti-mouse and anti-goat IgG (H + L) antibodies conjugated with Alexa Fluo 488 and Alex Fluo 568 (Molecular Probes, Eugene, Ore., USA, #SA5-10166, #SA5-10168, #A110556, #A21206). After being washed with PBS, the sections were incubated with the secondary antibody (1:500) for 90 min at room temperature and then rinsed in PBS. For nuclear staining, the tissue sections were incubated with 1:5,000 dilution of Hoechst (Sigma-Aldrich, USA, #23491-45-4) for 5 min at room temperature. After being washed, the tissue sections were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA). As a negative control, the primary antibody was replaced by 10% of mouse IgG (Vector Laboratories, USA, #I-2000). For cell double labeling, cells were fixed in cold acetone and methanol (1:1), permeabilized in 0.2% Triton X-100 at ice for 30 min and blocked with 10% donkey serum for 30 min at room temperature and two primary antibodies recognized by the donkey secondary antibody were incubated simultaneously overnight at 4 °C at the following dilutions: rabbit anti-FAK (1:100; Santa Cruz Biotechnology Inc., USA, #sc-557), goat anti-DSP (1:100; Santa Cruz Biotechnology Inc., USA, #sc-18328), mouse anti-Ocln (1:100; Life Technologies, USA, #33-150). After being washed, the cells were incubated with the secondary antibody conjugated with Alexa Fluo 486 green and Alexa Fluo 568 red (1:500; Molecular Probes, USA, #A21206, #SA5-10168) for 1 h at room temperature. For nuclear staining, the cells were incubated with 1:5,000 dilution of Hoechst (Sigma-Aldrich, USA, #23491-45-4) for 3 min at room temperature. After being washed, the cells were mounted in Vectashield mounting medium (Vector Laboratories, USA). As a negative control, the primary antibody was replaced by 10% mouse IgG (Vector Laboratories, USA, #I-2000). Images of Alexa Fluo 488 and Alex Fluo 568 staining of the various proteins were captured by a Nikon Eclipse TE2000S microscope with a filter by means of a digital cooled camera connected to a PC computer and analyzed with NIS-Elements 3.2 software. Images of nuclear staining with Hoechst were obtained via filter UV-2E/C, C86826. For each experiment, all slides were simultaneously processed for a specific antibody, so that homogeneity in the staining procedure was ensured between samples.
Effect of dentin sialoprotein on phosphorylation of occludin and FAK
Mouse dental papilla mesenchymal cells were treated with or without recombinant DSPf5 (2-24 μg/ml) in DMEM medium with 1% antibiotics at 37 °C in 5% CO2 atmosphere at given time periods. The cells were then washed with PBS and lysed with RIPA buffer (1 x PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mg/ml phenylmethylsulfonyl fluoride, 30 μl/ml aprotinin, 100 mM sodium orthovanadate; Santa Cruz Biotechnology Inc., USA). To characterize effect of DSPf5 on Ocln partners, Western blot assay was performed as described earlier16. Whole cell lysates were resolved by 7% SDS-PAGE gels and transferred to Trans-Blot membranes (Bio-Rad Laboratories, Inc., USA). The membrane was blocked with 5% non-fat milk in 1 x TBST buffer for 60 min at room temperature. After washing, the membranes were incubated with primary antibodies specific to Ocln (1:1,000, Life Technologies, USA, #33-150), p-Ocln-ser490 (1:500), FAK (1:1,000; Santa Cruz Biotechnology Inc., USA, #sc-557), p-FAK-ser722 (1:800, Santa Cruz Biotechnology Inc., USA, #sc-374668), p-FAK-tyr576 (1:1,000, Santa Cruz Biotechnology Inc., USA, #sc-16563-R), p-FAK-tyr-397 (1:1,000, Santa Cruz Biotechnology Inc., USA, #sc-11765-R), Akt (1:1,000, Santa Cruz Biotechnology Inc., USA, #sc-5298), p-Akt-ser473 (1:1,000, Santa Cruz Biotechnology Inc., USA, #sc-7985-R), and β-actin (1:1,500, Santa Cruz Biotechnology Inc., USA, #sc-1616) for overnight at 4 °C, respectively. The membranes were washed with 1 × TBST and incubated with diluted horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000-10,000, Pierce, USA) for 1 h at room temperature. After three washes, the membranes were detected using an enhanced chemiluminescence (ECL) kit (Millipore, USA). For block of effect of DSP on Ocln and FAK phosphorylation, the cells were treated with either DSPf5 or DSPf5 plus DSP antibody (8 or 16 μg/ml) or Ocln antibody (8 or 16 μg/ml) in DMEM medium with 1% antibiotics at 37 °C in 5% CO2 atmosphere for 1 h. The cells were harvested and phosphorylation of Ocln and FAK were carried out by Western blot assay.
Reparative dentin regeneration
Affi-Gel Blue (cross-linked agarose beads with covalently coupled Cibacron Blue F3-GA) was purchased from Bio-Rad laboratories Inc. and mixed with the DSP peptide (2 mg/ml). The mixture was centrifuged and pellet used for implantation. The experimental procedures were approved by the animal research committee of UTHSCSA. Twenty-five adult mice (C57BL/6) aged 1.5 months were anaesthetized by sodium pentobarbital (Sigma-Aldrich, USA). The first molars on the maxilla were cleaned by sterilized instruments. Exposed pulped cavities were prepared by ¼ diamond cylindrical burs with sterile saline cooling. The exposed pulped cavity on the left first molars was filled onto Affi-Gel blue gel as control and Affi-Gel blue gel with the DSP peptide on the right first molars. The cavities over the implanted materials were filled with Vitremer Glass Ionomer (GI) Core Build-Up/Restorative (Sku# 3303PEDO, 3 M ESPE, Dental Products, St. Paul, NM, USA) and sealed by light curing. Sealant well done in each animal was observed carefully. The animals were euthanatized at 1, 3, and 5 weeks after surgery. Tissue specimen were fixed in 4% paraformaldehyde at 4 °C overnight and embedded in paraffin wax after demineralization with 10% EDTA. The paraffin sections (5 μm in thickness) were morphologically examined after staining with hematoxylin and eosin (HE) (Agilent Technologies, USA).
Statistical analysis
Quantitative data were presented as means S.D. from three independent experiments and compared with the results of one-way ANOVA using GraphPad Prism 5 (GraphPad Software, Inc. La Jolla, CA, USA). The differences between groups were statistically significant at *p < 0.05 and **p < 0.01.
References
Erickson, C. A. & Reedy, M. V. Neural crest development: the interplay between morphogenesis and cell differentiation. Curr Top Dev Biol 40, 177–209 (1998).
Discher, D. E., Mooney, D. J. & Zandstra, P. W. Growth factors, matrices, and forces combine and control stem cells. Science 324, 1673–1677, doi:10.1126/science.1171643 (2009).
Morrison, S. J. & Spradling, A. C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611, doi:10.1016/j.cell.2008.01.038 (2008).
Bellahcene, A., Castronovo, V., Ogbureke, K. U., Fisher, L. W. & Fedarko, N. S. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): multifunctional proteins in cancer. Nat Rev Cancer 8, 212–226, doi:10.1038/nrc2345 (2008).
D’Souza, R. N. et al. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res 12, 2040–2049, doi:10.1359/jbmr.1997.12.12.2040 (1997).
Chen, S. et al. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J Biol Chem 280, 29717–29727, doi:10.1074/jbc.M502929200 (2005).
Qin, C., Brunn, J. C., Cadena, E., Ridall, A. & Butler, W. T. Dentin sialoprotein in bone and dentin sialophosphoprotein gene expressed by osteoblasts. Connect Tissue Res 44(Suppl 1), 179–183 (2003).
MacDougall, M. et al. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J Biol Chem 272, 835–842 (1997).
Wan, C. et al. The Dentin Sialoprotein (DSP) Domain Regulates Dental Mesenchymal Cell Differentiation through a Novel Surface Receptor. Scientific reports 6, 29666, doi:10.1038/srep29666 (2016).
Qin, C., Cook, R. G., Orkiszewski, R. S. & Butler, W. T. Identification and characterization of the carboxyl-terminal region of rat dentin sialoprotein. J Biol Chem 276, 904–909, doi:10.1074/jbc.M006271200 (2001).
Yamakoshi, Y. et al. Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J Biol Chem 281, 38235–38243, doi:10.1074/jbc.M607767200 (2006).
Ritchie, H. H. et al. Dentin sialoprotein (DSP) transcripts: developmentally-sustained expression in odontoblasts and transient expression in pre-ameloblasts. Eur J Oral Sci 105, 405–413 (1997).
Chen, S. et al. Expression and processing of small integrin-binding ligand N-linked glycoproteins in mouse odontoblastic cells. Arch Oral Biol 53, 879–889, doi:10.1016/j.archoralbio.2008.05.005 (2008).
Yuan, G., Yang, G., Song, G., Chen, Z. & Chen, S. Immunohistochemical localization of the NH(2)-terminal and COOH-terminal fragments of dentin sialoprotein in mouse teeth. Cell Tissue Res 349, 605–614, doi:10.1007/s00441-012-1418-4 (2012).
Yuan, G. et al. Dentin Sialoprotein is a Novel Substrate of Matrix Metalloproteinase 9 in vitro and in vivo. Scientific reports 7, 42449, doi:10.1038/srep42449 (2017).
Ozer, A. et al. Domain of dentine sialoprotein mediates proliferation and differentiation of human periodontal ligament stem cells. PLoS One 8, e81655, doi:10.1371/journal.pone.0081655 (2013).
Suzuki, S. et al. Dentin sialoprotein and dentin phosphoprotein have distinct roles in dentin mineralization. Matrix Biol 28, 221–229, doi:10.1016/j.matbio.2009.03.006 (2009).
Paine, M. L. et al. Dentin sialoprotein and dentin phosphoprotein overexpression during amelogenesis. J Biol Chem 280, 31991–31998, doi:10.1074/jbc.M502991200 (2005).
Xiao, S. et al. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet 27, 201–204, doi:10.1038/84848 (2001).
Kim, J. W. & Simmer, J. P. Hereditary dentin defects. J Dent Res 86, 392–399 (2007).
Rajpar, M. H. et al. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum Mol Genet 11, 2559–2565 (2002).
Tsukita, S., Furuse, M. & Itoh, M. Multifunctional strands in tight junctions. Nature reviews. Molecular cell biology 2, 285–293, doi:10.1038/35067088 (2001).
Jadlowiec, J. et al. Phosphophoryn regulates the gene expression and differentiation of NIH3T3, MC3T3-E1, and human mesenchymal stem cells via the integrin/MAPK signaling pathway. J Biol Chem 279, 53323–53330, doi:10.1074/jbc.M404934200 (2004).
Eapen, A., Ramachandran, A. & George, A. Dentin phosphoprotein (DPP) activates integrin-mediated anchorage-dependent signals in undifferentiated mesenchymal cells. J Biol Chem 287, 5211–5224, doi:10.1074/jbc.M111.290080 (2012).
Li, D. et al. Mutation identification of the DSPP in a Chinese family with DGI-II and an up-to-date bioinformatic analysis. Genomics 99, 220–226, doi:10.1016/j.ygeno.2012.01.006 (2012).
Lee, S. Y. et al. Effects of recombinant dentin sialoprotein in dental pulp cells. J Dent Res 91, 407–412, doi:10.1177/0022034511436113 (2012).
Furuse, M. et al. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol 123, 1777–1788 (1993).
Elmadih, A. et al. Natural Variation in Maternal Sensitivity Is Reflected in Maternal Brain Responses to Infant Stimuli. Behavioral neuroscience. doi:10.1037/bne0000161 (2016).
Dorfel, M. J. & Huber, O. Modulation of tight junction structure and function by kinases and phosphatases targeting occludin. J Biomed Biotechnol 2012, 807356, doi:10.1155/2012/807356 (2012).
Lacaz-Vieira, F., Jaeger, M. M., Farshori, P. & Kachar, B. Small synthetic peptides homologous to segments of the first external loop of occludin impair tight junction resealing. J Membr Biol 168, 289–297 (1999).
Ploss, A. et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457, 882–886, doi:10.1038/nature07684 (2009).
Cummins, P. M. Occludin: one protein, many forms. Mol Cell Biol 32, 242–250, doi:10.1128/MCB.06029-11 (2012).
LeBlanc, M. A. et al. A novel rearrangement of occludin causes brain calcification and renal dysfunction. Human genetics 132, 1223–1234, doi:10.1007/s00439-013-1327-y (2013).
Elsaid, M. F. et al. Whole genome sequencing identifies a novel occludin mutation in microcephaly with band-like calcification and polymicrogyria that extends the phenotypic spectrum. Am J Med Genet A 164A, 1614–1617, doi:10.1002/ajmg.a.36485 (2014).
Aggarwal, S., Bahal, A. & Dalal, A. Renal dysfunction in sibs with band like calcification with simplified gyration and polymicrogyria: Report of a new mutation and review of literature. European journal of medical genetics 59, 5–10, doi:10.1016/j.ejmg.2015.11.014 (2016).
Kitajiri, S. et al. Deafness in occludin-deficient mice with dislocation of tricellulin and progressive apoptosis of the hair cells. Biol Open 3, 759–766, doi:10.1242/bio.20147799 (2014).
Saitou, M. et al. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell 11, 4131–4142 (2000).
Saitou, M. et al. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. The Journal of cell biology 141, 397–408 (1998).
Hehlgans, S., Haase, M. & Cordes, N. Signalling via integrins: implications for cell survival and anticancer strategies. Biochim Biophys Acta 1775, 163–180, doi:10.1016/j.bbcan.2006.09.001 (2007).
Tilghman, R. W. & Parsons, J. T. Focal adhesion kinase as a regulator of cell tension in the progression of cancer. Semin Cancer Biol 18, 45–52, doi:10.1016/j.semcancer.2007.08.002 (2008).
Hakuno, D., Takahashi, T., Lammerding, J. & Lee, R. T. Focal adhesion kinase signaling regulates cardiogenesis of embryonic stem cells. J Biol Chem 280, 39534–39544, doi:10.1074/jbc.M505575200 (2005).
Tamura, Y. et al. Focal adhesion kinase activity is required for bone morphogenetic protein–Smad1 signaling and osteoblastic differentiation in murine MC3T3-E1 cells. J Bone Miner Res 16, 1772–1779, doi:10.1359/jbmr.2001.16.10.1772 (2001).
Ilic, D. et al. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377, 539–544, doi:10.1038/377539a0 (1995).
Sreenath, T. et al. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. The Journal of biological chemistry 278, 24874–24880, doi:10.1074/jbc.M303908200 (2003).
McKnight, D. A. & Fisher, L. W. Molecular evolution of dentin phosphoprotein among toothed and toothless animals. BMC Evol Biol 9, 299, doi:10.1186/1471-2148-9-299 (2009).
Iguchi, Y. et al. Intercellular junctions in odontoblasts of the rat incisor studied with freeze-fracture. Archives of oral biology 29, 487–497 (1984).
Sasaki, T. & Garant, P. R. Structure and organization of odontoblasts. The Anatomical record 245, 235–249, doi:10.1002/(sici)1097-0185(199606)245:2<235::aid-ar10>3.0.co;2-q (1996).
Arana-Chavez, V. E. & Katchburian, E. Freeze-fracture studies of the distal plasma membrane of rat odontoblasts during their differentiation and polarisation. European journal of oral sciences 106(Suppl 1), 132–136 (1998).
Hoshino, M. et al. Claudin rather than occludin is essential for differentiation in rat incisor odontoblasts. Oral diseases 14, 606–612, doi:10.1111/j.1601-0825.2007.01427.x (2008).
Munhoz, C. O. & Leblond, C. P. Deposition of calcium phosphate into dentin and enamel as shown by radioautography of sections of incisor teeth following injection of 45Ca into rats. Calcified tissue research 15, 221–235 (1974).
Nagai, N. & Frank, R. M. Electron microscopic autoradiography of Ca-45 during dentinogenesis. Cell and tissue research 155, 513–523 (1974).
Reith, E. J. The binding of calcium within the Golgi saccules of the rat odontoblast. The American journal of anatomy 147, 267–270, doi:10.1002/aja.1001470302 (1976).
Appleton, J. & Morris, D. C. An ultrastructural investigation of the role of the odontoblast in matrix calcification using the potassium pyroantimonate osmium method for calcium localization. Archives of oral biology 24, 467–475 (1979).
Tsukita, S., Furuse, M. & Itoh, M. Structural and signalling molecules come together at tight junctions. Current opinion in cell biology 11, 628–633 (1999).
Unda, F. J. et al. Dynamic assembly of tight junction-associated proteins ZO-1, ZO-2, ZO-3 and occludin during mouse tooth development. Histology and histopathology 18, 27–38 (2003).
Joao, S. M. & Arana-Chavez, V. E. Tight junctions in differentiating ameloblasts and odontoblasts differentially express ZO-1, occludin, and claudin-1 in early odontogenesis of rat molars. The anatomical record. Part A, Discoveries in molecular, cellular, and evolutionary biology 277, 338–343, doi:10.1002/ar.a.20021 (2004).
Inai, T., Sengoku, A., Hirose, E., Iida, H. & Shibata, Y. Differential expression of the tight junction proteins, claudin-1, claudin-4, occludin, ZO-1, and PAR3, in the ameloblasts of rat upper incisors. Anatomical record (Hoboken, N.J. : 2007) 291, 577–585, doi:10.1002/ar.20683 (2008).
Lee, T. Y. et al. Disruption of Nfic causes dissociation of odontoblasts by interfering with the formation of intercellular junctions and aberrant odontoblast differentiation. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society 57, 469–476, doi:10.1369/jhc.2009.952622 (2009).
Hata, M., Kawamoto, T., Kawai, M. & Yamamoto, T. Differential expression patterns of the tight junction-associated proteins occludin and claudins in secretory and mature ameloblasts in mouse incisor. Medical molecular morphology 43, 102–106, doi:10.1007/s00795-009-0482-7 (2010).
Lundgren, T., Nilsson, M., Ritchie, H. H. & Linde, A. Junctional proteins and Ca2+ transport in the rat odontoblast-like cell line MRPC-1. Calcified tissue international 68, 192–201 (2001).
Bleicher, F., Couble, M. L., Farges, J. C., Couble, P. & Magloire, H. Sequential expression of matrix protein genes in developing rat teeth. Matrix biology: journal of the International Society for Matrix Biology 18, 133–143 (1999).
Ikenouchi, J. et al. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. The Journal of cell biology 171, 939–945, doi:10.1083/jcb.200510043 (2005).
Murakami, T., Frey, T., Lin, C. & Antonetti, D. A. Protein kinase cbeta phosphorylates occludin regulating tight junction trafficking in vascular endothelial growth factor-induced permeability in vivo. Diabetes 61, 1573–1583, doi:10.2337/db11-1367 (2012).
Zhang, X. et al. DSPP mutation in dentinogenesis imperfecta Shields type II. Nature genetics 27, 151–152, doi:10.1038/84765 (2001).
Song, Y. et al. Phenotypes and genotypes in 2 DGI families with different DSPP mutations. Oral surgery, oral medicine, oral pathology, oral radiology, and endodontics 102, 360–374, doi:10.1016/j.tripleo.2005.06.020 (2006).
Gu, K., Chang, S., Ritchie, H. H., Clarkson, B. H. & Rutherford, R. B. Molecular cloning of a human dentin sialophosphoprotein gene. European journal of oral sciences 108, 35–42 (2000).
Siu, E. R., Wong, E. W., Mruk, D. D., Porto, C. S. & Cheng, C. Y. Focal adhesion kinase is a blood-testis barrier regulator. Proc Natl Acad Sci USA 106, 9298–9303, doi:10.1073/pnas.0813113106 (2009).
Kornberg, L., Earp, H. S., Parsons, J. T., Schaller, M. & Juliano, R. L. Cell adhesion or integrin clustering increases phosphorylation of a focal adhesion-associated tyrosine kinase. J Biol Chem 267, 23439–23442 (1992).
Burridge, K., Fath, K., Kelly, T., Nuckolls, G. & Turner, C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol 4, 487–525, doi:10.1146/annurev.cb.04.110188.002415 (1988).
Chiarugi, P. et al. Reactive oxygen species as essential mediators of cell adhesion: the oxidative inhibition of a FAK tyrosine phosphatase is required for cell adhesion. J Cell Biol 161, 933–944, doi:10.1083/jcb.200211118 (2003).
Zeng, L. et al. PTP alpha regulates integrin-stimulated FAK autophosphorylation and cytoskeletal rearrangement in cell spreading and migration. J Cell Biol 160, 137–146, doi:10.1083/jcb.200206049 (2003).
Zachary, I. Focal adhesion kinase. Int J Biochem Cell Biol 29, 929–934 (1997).
Grigera, P. R. et al. FAK phosphorylation sites mapped by mass spectrometry. J Cell Sci 118, 4931–4935, doi:10.1242/jcs.02696 (2005).
Chen, Z. et al. Klf10 regulates odontoblast differentiation and mineralization via promoting expression of dentin matrix protein 1 and dentin sialophosphoprotein genes. Cell and tissue research 363, 385–398, doi:10.1007/s00441-015-2260-2 (2016).
Wang, F. et al. Immortalized mouse dental papilla mesenchymal cells preserve odontoblastic phenotype and respond to bone morphogenetic protein 2. In Vitro Cell Dev Biol Anim 49, 626–637, doi:10.1007/s11626-013-9641-1 (2013).
Gronthos, S., Mankani, M., Brahim, J., Robey, P. G. & Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA 97, 13625–13630, doi:10.1073/pnas.240309797 (2000).
Xu, X., Chen, Z., Wang, Y., Yamada, Y. & Steffensen, B. Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and -2. Biochem J 392, 127–134, doi:10.1042/BJ20050650 (2005).
Acknowledgements
We thank Dr. David A. Antonetti for the anti-phosphorylated occludin antibody and Dr. Mikio Furuse for mouse occludin plasmid. This work has been generously supported by NIH Grant DE019802 from the NIDCR and by a pilot award of School of Dentistry, the University of Texas Health Science Center at San Antonio. We greatly acknowledge Dr. Dun-Xian Tan for his English grammar editing. The authors also appreciated three anonymous reviewers for their guidance with improving the quality and depth of our manuscript.
Author information
Authors and Affiliations
Contributions
Drs Wentong Li and Shuo Chen performed the main parts of this experiment. Dr. Lei Chen performed the initial plasmid constructions and GST-pull down assays. Drs Zhuo Chen and Feng Wang carried out immunohistochemistry. Drs Lian Wu and Junsheng Feng performed animal surgery and histological study. Drs Lisa Shoff and Xin Li participated in protein-protein interaction assay. Drs Kevin J. Donly, Mary MacDougall and Wentong Li analyzed and interpreted data. Drs Shuo Chen and Wentong Li wrote and revised this manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Li, W., Chen, L., Chen, Z. et al. Dentin sialoprotein facilitates dental mesenchymal cell differentiation and dentin formation. Sci Rep 7, 300 (2017). https://doi.org/10.1038/s41598-017-00339-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-00339-w
This article is cited by
-
Genome-wide screening for deubiquitinase subfamily identifies ubiquitin-specific protease 49 as a novel regulator of odontogenesis
Cell Death & Differentiation (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.