Abstract
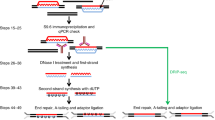
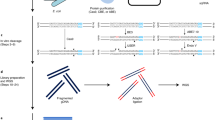
Nascent RNA may form a three-stranded structure with DNA, called an R-loop, which has been linked to fundamental biological processes such as transcription, replication and genome instability. Here, we provide a detailed protocol for a newly developed strategy, named R-ChIP, for robust capture of R-loops genome-wide. Distinct from R-loop-mapping methods based on the monoclonal antibody S9.6, which recognizes RNA–DNA hybrid structures, R-ChIP involves expression of an exogenous catalytically inactive RNASEH1 in cells to bind RNA–DNA hybrids but not resolve them. This is followed by chromatin immunoprecipitation (ChIP) of the tagged RNASEH1 and construction of a strand-specific library for deep sequencing. It takes ~3 weeks to establish a stable cell line expressing the mutant enzyme and 5 more days to proceed with the R-ChIP protocol. In principle, R-ChIP is applicable to both cell lines and animals, as long as the catalytically inactive RNASEH1 can be expressed to study the dynamics of R-loop formation and resolution, as well as its impact on the functionality of the genome. In our recent studies with R-ChIP, we showed an intimate spatiotemporal relationship between R-loops and RNA polymerase II pausing/pause release, as well as linking augmented R-loop formation to DNA damage response induced by driver mutations of key splicing factors associated with myelodysplastic syndrome (MDS).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
R-ChIP data for HEK293T and K562 cell lines are available at the NCBI GEO repository under the accession number GSE97072. All public datasets used in this study are indicated in the corresponding figure legends.
References
Aguilera, A. & Garcia-Muse, T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012).
Sanz, L. A. et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 (2016).
Wahba, L., Costantino, L., Tan, F. J., Zimmer, A. & Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30, 1327–1338 (2016).
Chen, L. et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell 68, 745–757 (2017).
Pefanis, E. et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015).
Graf, M. et al. Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell 170, 72–85 (2017).
Kabeche, L., Nguyen, H. D., Buisson, R. & Zou, L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359, 108–114 (2018).
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I. & Chedin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012).
Sugimoto, N. et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34, 11211–11216 (1995).
Duquette, M. L., Handa, P., Vincent, J. A., Taylor, A. F. & Maizels, N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 18, 1618–1629 (2004).
Roy, D., Yu, K. & Lieber, M. R. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 28, 50–60 (2008).
Roy, D. & Lieber, M. R. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol. 29, 3124–3133 (2009).
Roy, D., Zhang, Z., Lu, Z., Hsieh, C. L. & Lieber, M. R. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 30, 146–159 (2010).
Chedin, F. Nascent connections: R-loops and chromatin patterning. Trends Genet. 32, 828–838 (2016).
Stork, C. T. et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5, e17548 (2016).
Hamperl, S., Bocek, M. J., Saldivar, J. C., Swigut, T. & Cimprich, K. A. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170, 774–786 (2017).
Li, X. & Manley, J. L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378 (2005).
Garcia-Benitez, F., Gaillard, H. & Aguilera, A. Physical proximity of chromatin to nuclear pores prevents harmful R loop accumulation contributing to maintain genome stability. Proc. Natl. Acad. Sci. USA 114, 10942–10947 (2017).
Huertas, P. & Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003).
El Hage, A., French, S. L., Beyer, A. L. & Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 (2010).
Chang, E. Y. & Stirling, P. C. Replication fork protection factors controlling R-loop bypass and suppression. Genes (Basel) 8, 33 (2017).
Taneja, N. et al. SNF2 family protein Fft3 suppresses nucleosome turnover to promote epigenetic inheritance and proper replication. Mol. Cell 66, 50–62 (2017).
Chakraborty, P. & Grosse, F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.) 10, 654–665 (2011).
Cristini, A., Groh, M., Kristiansen, M. S. & Gromak, N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905 (2018).
Skourti-Stathaki, K., Proudfoot, N. J. & Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011).
Itoh, T. & Tomizawa, J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. USA 77, 2450–2454 (1980).
Xu, B. & Clayton, D. A. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 15, 3135–3143 (1996).
Sun, Q., Csorba, T., Skourti-Stathaki, K., Proudfoot, N. J. & Dean, C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621 (2013).
Boque-Sastre, R. et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA 112, 5785–5790 (2015).
Ohle, C. et al. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167, 1001–1013 (2016).
Belotserkovskii, B. P., Soo Shin, J. H. & Hanawalt, P. C. Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res. 45, 6589–6599 (2017).
Gan, W. et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25, 2041–2056 (2011).
Chen, L. et al. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol. Cell 69, 412–425.e6 (2018).
Costantino, L. & Koshland, D. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 34, 39–45 (2015).
Richard, P. & Manley, J. L. R loops and links to human disease. J. Mol. Biol. 429, 3168–3180 (2017).
Powell, W. T. et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl. Acad. Sci. USA 110, 13938–13943 (2013).
Nadel, J. et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin 8, 46 (2015).
Xu, W. et al. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 3, 704–714 (2017).
Cerritelli, S. M. & Crouch, R. J. Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics 53, 300–307 (1998).
Yu, K., Chedin, F., Hsieh, C. L., Wilson, T. E. & Lieber, M. R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4, 442–451 (2003).
Chen, P. B., Chen, H. V., Acharya, D., Rando, O. J. & Fazzio, T. G. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 22, 999–1007 (2015).
Dumelie, J. G. & Jaffrey, S. R. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. Elife 6, e28306 (2017).
Halasz, L. et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res. 27, 1063–1073 (2017).
Vanoosthuyse, V. Strengths and weaknesses of the current strategies to map and characterize R-loops. Noncoding RNA 4, E9 (2018).
Konig, F., Schubert, T. & Langst, G. The monoclonal S9.6 antibody exhibits highly variable binding affinities towards different R-loop sequences. PLoS ONE 12, e0178875 (2017).
Hartono, S. R. et al. The affinity of the S9.6 antibody for double-stranded RNAs impacts the accurate mapping of R-loops in fission yeast. J. Mol. Biol. 430, 272–284 (2018).
Phillips, D. D. et al. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 26, 376–381 (2013).
Li, X. et al. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 35, 940–950 (2017).
Bell, J. C. et al. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife 7, e27024 (2018).
Schmitz, K. M., Mayer, C., Postepska, A. & Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24, 2264–2269 (2010).
Bacolla, A., Wang, G. & Vasquez, K. M. New perspectives on DNA and RNA triplexes as effectors of biological activity. PLoS Genet. 11, e1005696 (2015).
Mondal, T. et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6, 7743 (2015).
O’Leary, V. B. et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 11, 474–485 (2015).
Postepska-Igielska, A. et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell 60, 626–636 (2015).
Li, Y., Syed, J. & Sugiyama, H. RNA-DNA triplex formation by long noncoding RNAs. Cell Chem. Biol. 23, 1325–1333 (2016).
Nowotny, M. et al. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 27, 1172–1181 (2008).
Nguyen, H. D. et al. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell 65, 832–847 (2017).
Bhatia, V. et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365 (2014).
Legros, P., Malapert, A., Niinuma, S., Bernard, P. & Vanoosthuyse, V. RNA processing factors Swd2.2 and Sen1 antagonize RNA Pol III-dependent transcription and the localization of condensin at Pol III genes. PLoS Genet. 10, e1004794 (2014).
Tresini, M. et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58 (2015).
Rhee, H. S. & Pugh, B. F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011).
Rotem, A. et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33, 1165–1172 (2015).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Core, L. J. et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320 (2014).
Acknowledgements
The authors are grateful to members of the Fu lab for cooperation, reagent sharing and insightful discussion that improved this work. We thank T. Hishida of the J.C. Izpisua Belmonte lab (Gene Expression Laboratories, Salk Institute for Biological Studies) for providing the pPyCAG plasmid and W. Li for critical comments on the manuscript. This work was supported by NIH grants (GM049369, GM052872, HG004659 and DK098808) to X.-D.F. and Start-up funds of Wuhan University (1304/413100052 and 1304/413100072) to L.C. We thank S. Dowdy (University of California, San Diego) for providing HEK293T cells
Author information
Authors and Affiliations
Contributions
L.C. and X.-D.F. conceived the idea; L.C. and J.-Y.C. codeveloped the R-ChIP method and data analysis pipeline; L.C. and X.Z. performed the experiments. J.-Y.C. performed bioinformatics analysis with assistance from L.C.; L.C., J.-Y.C. and X.-D.F. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Chen, L. et al. Mol. Cell 68, 745–757 (2017): https://doi.org/10.1016/j.molcel.2017.10.008
Chen, L. et al. Mol. Cell 69, 412–425.e6 (2018): https://doi.org/10.1016/j.molcel.2017.12.029
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, JY., Zhang, X., Fu, XD. et al. R-ChIP for genome-wide mapping of R-loops by using catalytically inactive RNASEH1. Nat Protoc 14, 1661–1685 (2019). https://doi.org/10.1038/s41596-019-0154-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0154-6
This article is cited by
-
The chromatin-associated RNAs in gene regulation and cancer
Molecular Cancer (2023)
-
High-throughput techniques enable advances in the roles of DNA and RNA secondary structures in transcriptional and post-transcriptional gene regulation
Genome Biology (2022)
-
Structural basis of R-loop recognition by the S9.6 monoclonal antibody
Nature Communications (2022)
-
Sources, resolution and physiological relevance of R-loops and RNA–DNA hybrids
Nature Reviews Molecular Cell Biology (2022)
-
ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.