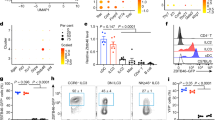
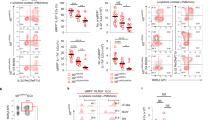
Abstract
Innate lymphoid cells (ILCs) are tissue-resident lymphocytes categorized on the basis of their core regulatory programs and the expression of signature cytokines. Human ILC3s that produce the cytokine interleukin-22 convert into ILC1-like cells that produce interferon-γ in vitro, but whether this conversion occurs in vivo remains unclear. In the present study we found that ILC3s and ILC1s in human tonsils represented the ends of a spectrum that included additional discrete subsets. RNA velocity analysis identified an intermediate ILC3–ILC1 cluster, which had strong directionality toward ILC1s. In humanized mice, the acquisition of ILC1 features by ILC3s showed tissue dependency. Chromatin studies indicated that the transcription factors Aiolos and T-bet cooperated to repress regulatory elements active in ILC3s. A transitional ILC3–ILC1 population was also detected in the human intestine. We conclude that ILC3s undergo conversion into ILC1-like cells in human tissues in vivo, and that tissue factors and Aiolos were required for this process.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Microarray data, bulk RNA sequencing and chromatin profiling data have been deposited in the GEO repository under accession code GSE130775. All the other data will be available upon reasonable request.
Change history
06 August 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Diefenbach, A., Colonna, M. & Koyasu, S. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365 (2014).
Eberl, G., Colonna, M., Di Santo, J. P. & McKenzie, A. N. Innate lymphoid cells: a new paradigm in immunology. Science 348, aaa6566 (2015).
Klose, C. S. & Artis, D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17, 765–774 (2016).
Ebbo, M., Crinier, A., Vely, F. & Vivier, E. Innate lymphoid cells: major players in inflammatory diseases. Nat. Rev. Immunol. 17, 665–678 (2017).
Huang, Q., Seillet, C. & Belz, G. T. Shaping innate lymphoid cell diversity. Front. Immunol. 8, 1569 (2017).
Bjorklund, A. K. et al. The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17, 451–460 (2016).
Gury-BenAri, M. et al. The Spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246 e1213 (2016).
Colonna, M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity 48, 1104–1117 (2018).
DuPage, M. & Bluestone, J. A. Harnessing the plasticity of CD4+ T cells to treat immune-mediated disease. Nat. Rev. Immunol. 16, 149–163 (2016).
Johnson, J. L. & Vahedi, G. Do memory CD4 T cells keep their cell-type programming: plasticity versus fate commitment? Epigenome: a dynamic vehicle for transmitting and recording cytokine signaling. Cold Spring Harb. Perspect. Biol. 10, a028779 (2018).
Cella, M., Otero, K. & Colonna, M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc. Natl Acad. Sci. USA 107, 10961–10966 (2010).
Montaldo, E., Juelke, K. & Romagnani, C. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur. J. Immunol. 45, 2171–2182 (2015).
Bernink, J. H. et al. Interleukin-12 and -23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat+ innate lymphocytes. Immunity 33, 736–751 (2010).
Sciume, G. et al. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209, 2331–2338 (2012).
Klose, C. S. et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature 494, 261–265 (2013).
Rankin, L. C. et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the notch pathway. Nat. Immunol. 14, 389–395 (2013).
Lee, J. S. et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13, 144–151 (2011).
Viant, C. et al. Transforming growth factor-beta and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 9, ra46 (2016).
Chea, S. et al. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci. Signal. 9, ra45 (2016).
Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity 38, 769–781 (2013).
Koues, O. I. et al. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell 165, 1134–1146 (2016).
Zhang, J. et al. S100A4 contributes to colitis development by increasing the adherence of Citrobacter rodentium in intestinal epithelial cells. Sci. Rep. 7, 12099 (2017).
Amir el, A. D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Holmes, T. D. & Bryceson, Y. T. Natural killer cell memory in context. Semin. Immunol. 28, 368–376 (2016).
Freud, A. G., Mundy-Bosse, B. L., Yu, J. & Caligiuri, M. A. The broad spectrum of human natural killer cell diversity. Immunity 47, 820–833 (2017).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Rongvaux, A. et al. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol. 32, 364–372 (2014).
Herndler-Brandstetter, D. et al. Humanized mouse model supports development, function, and tissue residency of human natural killer cells. Proc. Natl Acad. Sci. USA 114, E9626–E9634 (2017).
Kioussis, D. Aiolos: an ungrateful member of the Ikaros family. Immunity 26, 275–277 (2007).
Petzold, G., Fischer, E. S. & Thoma, N. H. Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature 532, 127–130 (2016).
Ma, S. et al. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol. Cell. Biol. 30, 4149–4158 (2010).
Allan, D. S. et al. An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol. 8, 340–351 (2015).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Koipally, J., Renold, A., Kim, J. & Georgopoulos, K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18, 3090–3100 (1999).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Holmes, M. L. et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J. 33, 2721–2734 (2014).
Mazzurana, L. et al. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3−ILC1/NK cell transdifferentiation. Eur. J. Immunol. (in the press).
Mikami, Y. et al. NCR+ ILC3 maintain larger STAT4 reservoir via T-BET to regulate type 1 features upon IL-23 stimulation in mice. Eur. J. Immunol. 48, 1174–1180 (2018).
Quintana, F. J. et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat. Immunol. 13, 770–777 (2012).
Simoni, Y. & Newell, E. W. Toward meaningful definitions of innate-lymphoid-cell subsets. Immunity 46, 760–761 (2017).
Bernink, J. H., Mjosberg, J. & Spits, H. Human ILC1: to be or not to be. Immunity 46, 756–757 (2017).
Allan, D. S. J. et al. Transcriptome analysis reveals similarities between human blood CD3– CD56(bright) cells and mouse CD127+ innate lymphoid cells. Sci. Rep. 7, 3501 (2017).
Simoni, Y. et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity 48, 1060 (2018).
Zhou, L. et al. Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409 (2019).
Jameson, S. C. & Masopust, D. Understanding subset diversity in T cell memory. Immunity 48, 214–226 (2018).
Robinette, M. L. et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015).
Bando, J. K. et al. The tumor necrosis factor superfamily member RANKL suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity 48, 1208–1219 (2018).
Zerbino, D. R. et al. Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Acknowledgements
We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support grant no. P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA grant no. UL1 TR000448 from the National Center for Research Resources. We thank E. Lantelme and D. Brinja, and the Pathology and Immunology Flow Cytometry Core for cell sorting. We thank O. Malkova, R. Lin and S. Oh, and the Center for Human Immunology at Washington University for help in processing the samples for mass spectrometry and for advice on analysis. We thank the Washington University Digestive Disease Research Core (NIDDK P30 DK052574) for support. We thank Regeneron for providing MISTRG-6 and IL-15 humanized mice. We thank S. Henikoff (Fred Hutchison Cancer Research Center, Seattle, USA) for providing protein-A-Mnase. This work was in part supported by grant nos UO1 AI095542 and RO1 DE025884 (to M. Colonna) and RO1 AI134035 (to M. Colonna and E.M.O.). R.A.F. is supported by the Howard Hughes Institute.
Author information
Authors and Affiliations
Contributions
C.S, P.L.C. and S.Z. contributed equally to this work. M. Cella designed, performed and interpreted experiments. R.G. and S.Z. analyzed the scRNA-seq data and wrote the methods for the scRNA-seq analysis. C.S. generated Aiolos- and T-bet-transduced MNK3 cells. M.L.R. and V.P. analyzed the microarray data and RNA-seq data. K.Z. and M.N.A. provided bioinformatic support. J.K.B., K.Y. and V.C. helped in flow cytometry data presentation and analysis. C.F. and R.F. generated libraries for scRNA-seq. J.S. provided critical advice for Cytof analysis. W.G., L.-L.L. and M.B. provided critical insights to the study. S.G., R.A.F. and L.S. provided key reagents. P.L.C. performed the cut-and-run experiment and interpreted data under the supervision of E.M.O. S.A.J. and M. Colonna supervised the study. M. Cella, S.A.J. and M. Colonna wrote the manuscript and all the authors contributed to editing and suggestions.
Corresponding authors
Ethics declarations
Competing interests
R.G., S.Z., W.G., J.S., L.-L.L., M.B. and S.A.J. are Pfizer employees. M. Colonna received funding from Pfizer to study ILC biology in inflammatory bowel disease. The other authors declare no conflicts of interests.
Additional information
Peer review information: I. Visan was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Representative gating strategy for sorting ILC subsets.
a-e, Dot plots illustrating gatings. Gates were applied on lymphocytes (a), singlets (b), CD56+CD3–CD19 (c), NKp44+CD103– (d) to identify CD300LF+CCR6+ ILC3a (e), or NKp44+CD103+ (d) cells to identify ILC3b, ILC1b and ILC1a (f). One representative experiment of more than 50 is shown.
Supplementary Figure 2 Bulk RNA-seq of ILC1a, ILC1b, ILC3b, and ILC3a subsets.
a, Volcano plot of differentially expressed (DE) genes between ILC1a (blue) and ILC3a (red) subsets (p < 0.05, Base Mean > 50). b, Selected transcription factors, effector molecules, and cell surface proteins that are differentially expressed between ILC3a and ILC1a (p < 0.05, Wald test). c, Heatmap of normalized expression, z-scored by row, of all differentially expressed genes (p< 0.05) between ILC1a/ILC1b and ILC3a/ILC3b. Green boxes indicate clusters of genes which form a gradient of expression between ILC1a and ILC3a. d, Venn diagram of ILC subset-signature genes. ILC1a and ILC3a signature genes were determined by selecting genes enriched in ILC1a (FC > 0, p <0.05) and ILC3a (FC < 0, p <0.05) when comparing ILC1a and ILC3a. ILC1b signature genes were determined by selecting genes enriched in ILC1b (FC < 0, p <0.05) when comparing DE genes between ILC1a and ILC1b. ILC3b signature genes were determined by selecting genes enriched in ILC3b (FC < 0, p <0.05) when comparing DE genes between ILC3a and ILC3b. Data are obtained from 3 independent biological replicates each subset.
Supplementary Figure 3 ScRNAseq of the ILC3-ILC1 spectrum identifies an intermediate cluster transitioning to ILC1s.
a, Unsupervised tSNE analysis of ILC3-ILC1 subsets to define clusters. b, Heat map showing the top 10 differentially expressed genes in each cluster. Top genes expressed in each cluster are indicated on the right of the heat map. c, tSNE of representative ILC3-related or ILC1-related genes. d, RNA velocity analysis to predict cell dynamics. One donor representative of two is shown.
Supplementary Figure 4 Cytokine production in clones derived from ILC3a, ILC3b, ILC1b and ILC3a.
a, IFN-γ and IL-22 production by 2 of the most represented clones (panels on the left and in the middle) and by one of the least represented clones (panels on the right) derived from ILC3a, ILC3b, ILC1b and ILC1a. i, Percentages of cells producing IL-22 only, IFN-γ plus IL-22, or IFN-γ only in each clone tested. Data are mean±SD. A total of 98 clones were tested (ILC3a=33; ILC3b=11; ILC1b=29; ILC1a=25). One donor representative of 2 is shown. Significance was calculated by ordinary one-way ANOVA multiple comparison test using the Prism7 software. *p≤0.05, **p≤0.001, ***p≤0.005, ****p≤0.0001.
Supplementary Figure 5 In vivo transfer of human ILC3a into humanized mice.
a, Schematic design of in vivo transfer of ILC3a into MISTRG-6-15 mice. b, Identification of human cells in spleen, liver, intra-epithelia lymphocytes (IEL) and lamina propria (LP) of MISTRG-6-15 mice 4 weeks after transfer by staining for human CD45 and human MHCI. c, Percentages of human ILC3s in spleen and liver (n=5). Data are mean±SD. Significance was calculated by unpaired t test using the Prism7 software. ***p≤0.005.
Supplementary Figure 6 Functional analysis of in vivo transferred ILC3a into humanized mice.
a, Expression of RORγt, Aiolos and T-bet in ILC3a at the time of transfer. b, Production of IL-22 and IFN-γ by ILC3a at the time of transfer in the absence or presence of IL-23 stimulation. c, Percentages of human cells present in mouse spleen and liver at day 19 or day 39 post-transfer. d, Expression of RORγt, Aiolos and T-bet in ILCs from spleen and liver at day 19 and day 39 post-transfer in one representative mouse. e-f, Quantification of the percentages of Aiolos+ (e), Tbet+ (f) at the indicated time points in spleen and liver (n=4 mice, each time point). g, Production of IFN-γ and IL-22 by ILCs from spleen and liver at day 19 and day 39 post transfer in one representative mouse. Cells were stimulated with PMA/I. h-i, Quantification of the percentages of IFN-γ producing (h) and IL-22-secreting (i) cells at the indicated time points (n=4 mice, each time point) in spleen and liver. One experiment representative of two is shown. e, f, h, i, Data are mean±SD. Significance was calculated by ordinary one-way ANOVA multiple comparison test using the Prism7 software. *p≤0.05, **p≤0.001, ****p≤0.0001. Spl, spleen; Lv, liver.
Supplementary Figure 7 Expression of T-bet and Aiolos in transduced MNK3 cells.
a, Expression of the reporter proteins GFP and mCherry indicative of T-bet and Aiolos in untransduced MNK3 cells (control) and MNK3 transduced with T-bet, or both T-bet and Aiolos. b, Expression of the transduced transcription factors by intracellular staining. One representative of 3 experiment is shown. c, Intracellular staining for IL-22 in control MNK3 cells or MNK3 transduced with Aiolos and stimulated with IL-23 and IL-1β. d, IL-22 in supernatants of stimulated cells assessed by ELISA (n=5). Data represent mean±SD. One experiment representative of 4 (c) or 2 (d) is shown.
Supplementary Figure 8 Combined expression of Aiolos and T-bet represses type 3 signature genes.
a, UCSC snapshots of the Rorc locus. Tracks represent cut and run for H3K27ac (green, 0-300 RPKM), IKZF3 (blue, 0 to 100 RPKM) or ATAC-seq (red, 0 to 100 RPKM). Fold change plots below H3K27ac tracks represent differences between H3K27ac in MNK3 expressing T-bet or both Aiolos and T-bet. Conserved IKZF3 motifs between mouse loci (top snapshots) and human loci (bottom snapshots) are indicated in dashed lines. Relative kb scale and gene locations are indicated on top. b, RORγt expression by intracellular staining in control MNK3, MNK3 expressing T-bet and MNK3 expressing Aiolos and T-bet. c, RORγt MFI (n=3). Significance was calculated by ordinary one-way ANOVA multiple comparison test using the Prism7 software. *p≤0.05. One experiment representative of 3 is shown. d, UCSC snapshots of representative ILC1/NK genes (Ifng, Eomes), housekeeping genes (B2m, Suv39h2) and ILC3 genes (Il17a/f, S100a4). Tracks represent cut and run for H3K27ac (green, 0-300 RPKM) or IKZF3 (blue, 0 to 100 RPKM). Relative kb scale and gene locations are indicated on top. e, Specificity of Aiolos binding peaks at the II22 locus. Peaks are not observed on control or Tbet transduced MNK3. f, Mean H3K27ac cut and run values across all H3K27ac peaks that overlap (left) or do not overlap (right) IKZF3 peaks. g, De-novo motif prediction of genome-wide IKZF3 peaks. Left: de novo peak. Middle: closest match in the HOMER database. Right: p-value of motif enrichment. Rep, replicate.
Supplementary information
Supplementary Information
Supplementary Figures 1-8
Rights and permissions
About this article
Cite this article
Cella, M., Gamini, R., Sécca, C. et al. Subsets of ILC3−ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat Immunol 20, 980–991 (2019). https://doi.org/10.1038/s41590-019-0425-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0425-y
This article is cited by
-
The transcription factor Aiolos restrains the activation of intestinal intraepithelial lymphocytes
Nature Immunology (2024)
-
Group 3 innate lymphoid cells in intestinal health and disease
Nature Reviews Gastroenterology & Hepatology (2024)
-
Multiscale 3D genome organization underlies ILC2 ontogenesis and allergic airway inflammation
Nature Immunology (2023)
-
Proline uptake promotes activation of lymphoid tissue inducer cells to maintain gut homeostasis
Nature Metabolism (2023)
-
Innate lymphoid cells and innate-like T cells in cancer — at the crossroads of innate and adaptive immunity
Nature Reviews Cancer (2023)