Abstract
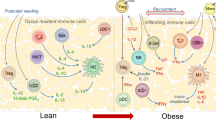
γδ T cells are situated at barrier sites and guard the body from infection and damage. However, little is known about their roles outside of host defense in nonbarrier tissues. Here, we characterize a highly enriched tissue-resident population of γδ T cells in adipose tissue that regulate age-dependent regulatory T cell (Treg) expansion and control core body temperature in response to environmental fluctuations. Mechanistically, innate PLZF+ γδ T cells produced tumor necrosis factor and interleukin (IL) 17 A and determined PDGFRα+ and Pdpn+ stromal-cell production of IL-33 in adipose tissue. Mice lacking γδ T cells or IL-17A exhibited decreases in both ST2+ Treg cells and IL-33 abundance in visceral adipose tissue. Remarkably, these mice also lacked the ability to regulate core body temperature at thermoneutrality and after cold challenge. Together, these findings uncover important physiological roles for resident γδ T cells in adipose tissue immune homeostasis and body-temperature control.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
07 February 2019
In the version of this article initially published, three authors (Hui-Fern Kuoy, Adam P. Uldrich and Dale. I. Godfrey) and their affiliations, acknowledgments and contributions were not included. The correct information is as follows:
Ayano C. Kohlgruber1,2, Shani T. Gal-Oz3, Nelson M. LaMarche1,2, Moto Shimazaki1, Danielle Duquette4, Hui-Fern Koay5,6, Hung N. Nguyen1, Amir I. Mina4, Tyler Paras1, Ali Tavakkoli7, Ulrich von Andrian2,8, Adam P. Uldrich5,6, Dale I. Godfrey5,6, Alexander S. Banks4, Tal Shay3, Michael B. Brenner1,10* and Lydia Lynch1,4,9,10*
1Division of Rheumatology, Immunology and Allergy, Brigham and Women’s Hospital, Boston, MA, USA. 2Division of Medical Sciences, Harvard Medical School, Boston, MA, USA. 3Department of Life Sciences, Ben-Gurion University of the Negev, Beersheba, Israel. 4Division of Endocrinology, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA. 5Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Parkville, Australia. 6ARC Centre of Excellence in Advanced Molecular Imaging, University of Melbourne, Parkville, Australia. 7Department of General and Gastrointestinal Surgery, Brigham and Women’s Hospital, Boston, MA, USA. 8Department of Microbiology and Immunology, Harvard Medical School, Boston, MA, USA. 9School of Biochemistry and Immunology, Trinity College, Dublin, Ireland. 10These authors jointly supervised this work: Michael B. Brenner, Lydia Lynch. *e-mail: mbrenner@research.bwh.harvard.edu; llynch@bwh.harvard.edu
Acknowledgements
We thank A.T. Chicoine, flow cytometry core manager at the Human Immunology Center at BWH, for flow cytometry sorting. We thank D. Sant’Angelo (Rutgers Cancer Institute) for providing Zbtb16–/– mice and R. O’Brien (National Jewish Health) for providing Vg4/6–/– mice. Supported by NIH grant R01 AI11304603 (to M.B.B.), ERC Starting Grant 679173 (to L.L.), the National Health and Medical Research Council of Australia (1013667), an Australian Research Council Future Fellowship (FT140100278 for A.P.U.) and a National Health and Medical Research Council of Australia Senior Principal Research Fellowship (1117766 for D.I.G.).
Author contributions
A.C.K., L.L., and M.B.B. conceived and designed the experiments, and wrote the manuscript. A.C.K., N.M.L., L.L., H.N.N., M.S., T.P., and D.D. performed the experiments. S.T.G.-O. and T.S. performed the RNA-seq analysis. A.S.B. and A.I.M. provided advice and performed the CLAMS experiments. A.T. provided human bariatric patient samples. Parabiosis experiments were performed in the laboratory of U.v.A. H.-F.K., A.P.U. and D.I.G provided critical insight into the TCR chain usage of PLZF+ γδ T cells. M.B.B., N.M.L., and L.L. critically reviewed the manuscript.
The errors have been corrected in the HTML and PDF version of the article.
Correction to: Nature Immunology doi:10.1038/s41590-018-0094-2 (2018), published online 18 April 2018.
References
Brestoff, J. R. & Artis, D. Immune regulation of metabolic homeostasis in health and disease. Cell 161, 146–160 (2015).
Kohlgruber, A. & Lynch, L. Adipose tissue inflammation in the pathogenesis of type 2 diabetes. Curr. Diab. Rep. 15, 92 (2015).
Lynch, L. et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of Treg cells and macrophages in adipose tissue. Nat. Immunol. 16, 85–95 (2015).
Lynch, L. et al. Adipose tissue invariant NKT cells protect against diet-induced obesity and metabolic disorder through regulatory cytokine production. Immunity 37, 574–587 (2012).
Molofsky, A. B. et al. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210, 535–549 (2013).
Nussbaum, J. C. et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013).
Lynch, L. et al. iNKT cells induce FGF21 for thermogenesis and are required for maximal weight loss in GLP1 therapy. Cell Metab. 24, 510–519 (2016).
Brestoff, J. R. et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature 519, 242–246 (2015).
Feuerer, M. et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15, 930–939 (2009).
Cipolletta, D. et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012).
Vasanthakumar, A. et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol. 16, 276–285 (2015).
Bapat, S. P. et al. Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature 528, 137–141 (2015).
Cipolletta, D. Adipose tissue-resident regulatory T cells: phenotypic specialization, functions and therapeutic potential. Immunology 142, 517–525 (2014).
Panduro, M., Benoist, C. & Mathis, D. Tissue Tregs. Annu. Rev. Immunol. 34, 609–633 (2016).
Kolodin, D. et al. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 21, 543–557 (2015).
Molofsky, A. B. et al. Interleukin-33 and interferon-γ counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity 43, 161–174 (2015).
Mathis, D. IL-33, imprimatur of adipocyte thermogenesis. Cell 166, 794–795 (2016).
Odegaard, J. I. et al. Perinatal licensing of thermogenesis by IL-33 and ST2. Cell 166, 841–854 (2016).
Lee, M.-W. et al. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 160, 74–87 (2015).
Jackson-Jones, L. H. et al. Fat-associated lymphoid clusters control local IgM secretion during pleural infection and lung inflammation. Nat. Commun. 7, 12651 (2016).
Pichery, M. et al. Endogenous IL-33 is highly expressed in mouse epithelial barrier tissues, lymphoid organs, brain, embryos, and inflamed tissues: in situ analysis using a novel Il-33-LacZ gene trap reporter strain. J. Immunol. 188, 3488–3495 (2012).
Paget, C. et al. CD3bright signals on γδ T cells identify IL-17A-producing Vγ6Vδ1+ T cells. Immunol. Cell Biol. 93, 198–212 (2015).
Ribot, J. C. et al. CD27 is a thymic determinant of the balance between interferon-γ- and interleukin 17-producing γδ T cell subsets. Nat. Immunol. 10, 427–436 (2009).
Kreslavsky, T. et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. Proc. Natl Acad. Sci. USA 106, 12453–12458 (2009).
Lu, Y., Cao, X., Zhang, X. & Kovalovsky, D. PLZF controls the development of fetal-derived IL-17+Vγ6+ γδ T cells. J. Immunol. 195, 4273–4281 (2015).
Savage, A. K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008).
Alonzo, E. S. & Sant’Angelo, D. B. Development of PLZF-expressing innate T cells. Curr. Opin. Immunol. 23, 220–227 (2011).
Vantourout, P. & Hayday, A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 13, 88–100 (2013).
Wei, Y.-L. et al. A highly focused antigen receptor repertoire characterizes γδ T cells that are poised to make IL-17 rapidly in naive animals. Front. Immunol. 6, 118 (2015).
Roark, C. L. et al. Subset-specific, uniform activation among Vγ6/Vδ1+ γδ T cells elicited by inflammation. J. Leukoc. Biol. 75, 68–75 (2004).
Van Dyken, S. J. et al. Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and γδ T cells. Immunity 40, 414–424 (2014).
Vannella, K. M. et al. Combinatorial targeting of TSLP, IL-25, and IL-33 in type 2 cytokine-driven inflammation and fibrosis. Sci. Transl. Med. 8, 337ra65 (2016).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014).
Kuswanto, W. et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity 44, 355–367 (2016).
Hepworth, M. R. Innate lymphoid cell regulation: meeting the long-lost cousin. Trends Immunol. 38, 873–874 (2017).
Zúñiga, L. A. et al. IL-17 regulates adipogenesis, glucose homeostasis, and obesity. J. Immunol. 185, 6947–6959 (2010).
Wood, I. S., Wang, B. & Trayhurn, P. IL-33, a recently identified interleukin-1 gene family member, is expressed in human adipocytes. Biochem. Biophys. Res. Commun. 384, 105–109 (2009).
Zeyda, M. et al. Severe obesity increases adipose tissue expression of interleukin-33 and its receptor ST2, both predominantly detectable in endothelial cells of human adipose tissue. Int. J. Obes. (Lond.) 37, 658–665 (2013).
Martin, N. T. & Martin, M. U. Interleukin 33 is a guardian of barriers and a local alarmin. Nat. Immunol. 17, 122–131 (2016).
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 (2008).
Wagers, A. J., Sherwood, R. I., Christensen, J. L. & Weissman, I. L. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science 297, 2256–2259 (2002).
Okada, K. et al. Thioesterase superfamily member 1 suppresses cold thermogenesis by limiting the oxidation of lipid droplet-derived fatty acids in brown adipose tissue. Mol. Metab. 5, 340–351 (2016).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Acknowledgements
We thank A.T. Chicoine, flow cytometry core manager at the Human Immunology Center at BWH, for flow cytometry sorting. We thank D. Sant’Angelo (Rutgers Cancer Institute) for providing Zbtb16–/– mice and R. O’Brien (National Jewish Health) for providing Vg4/6–/–- mice. Supported by NIH grant R01 AI11304603 (to M.B.B.), ERC Starting Grant 679173 (to L.L.), the National Health and Medical Research Council of Australia (1013667), an Australian Research Council Future Fellowship (FT140100278 for A.P.U.) and a National Health and Medical Research Council of Australia Senior Principal Research Fellowship (1117766 for D.I.G.).
Author information
Authors and Affiliations
Contributions
A.C.K., L.L., and M.B.B. conceived and designed the experiments, and wrote the manuscript. A.C.K., N.M.L., L.L., H.N.N., M.S., T.P., and D.D. performed the experiments. S.T.G.-O. and T.S. performed the RNA-seq analysis. A.S.B. and A.I.M. provided advice and performed the CLAMS experiments. A.T. provided human bariatric patient samples. Parabiosis experiments were performed in the laboratory of U.v.A. H.-F.K., A.P.U. and D.I.G provided critical insight into the TCR chain usage of PLZF+ γδ T cells. M.B.B., N.M.L., and L.L. critically reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.B.B. is a consultant to Roche.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Immunophenotyping panels for adipose immune-cell quantification.
(a) Representative flow cytometry plots to identify ILC2s, γδ T, CD4+ T, Foxp3+ Treg, and ST2+Foxp3+ Treg cells. (b) Representative flow cytometry plots to identify eosinophils, B220+CD19+ B, CD19+ B, NK, iNKT, and CD8+ T cells. (c) Numbers of CD4+ T, CD8+ T, eosinophils, CD19+ B, B220+CD19+ B, and NK cells per gram of eWAT at 5, 8, 11, 21 and 28 wks of age in male mice (n = 5, pooled). Each symbol represents an individual mouse; small horizontal lines indicate the mean. Data are representative across two experiments (a,b,c; mean ± s.e.m. in c).
Supplementary Figure 2 ILC2, iNKT, and Treg numbers in IL-17A-knockout and Vγ4/6-knockout mice.
(a) Numbers (left) and frequency (right) of ILC2s in eWAT from WT, Vg4/6–/– and Il17a–/– 16 wk old mice (n = 5, pooled). (b) Numbers (left) and frequency (right) of iNKTs in eWAT from WT, Vg4/6–/– and Il17a–/– 16 wk old mice (n = 5, pooled). (c) Quantification of numbers (top) and frequencies (bottom) of Treg cells and ST2+ Treg cells from spleen, lung, and adipose tissue from WT, Vg4/6–/– and Il17a–/– 16 wk old mice (n ≥ 3). (d) IL-33 protein from SVF eWAT lysates of 11 wk male WT and Il17a–/– mice normalized to total SVF protein by ELISA (n ≥ 3). (e) Numbers (top) and frequency (bottom) of Treg cells and ST2+ Treg cells from WT and Il17a–/– eWAT at 11 wks of age (n ≥ 4). Each symbol represents an individual mouse; small horizontal lines indicate the mean. NS, not significant (P > 0.05); * P < 0.05; **** P < 0.0001 (One-way ANOVA in a-c; Student’s t test in d-e). Data are pooled across two experiments (a-e; mean ± s.e.m. in a-e).
Supplementary Figure 3 In vitro and in vivo cytokine stimulations of epididymal adipose stromal cells.
(a) 3T3L1 adipose fibroblasts were unstimulated (unstim) or stimulated with TNFlo (0.1ng/mL), TNFhi (1ng/mL), IL-17Alo (0.1ng/mL), IL-17Ahi (1ng/mL), IL-1βlo (0.1ng/mL), IL-1βhi (1ng/mL), IFN-γlo (0.1ng/mL), IFN-γhi (1ng/mL), or a combination of the cytokines as indicated for 18h. IL-33 protein was measured by ELISA. (b) WT mice were injected with saline or TNF (1 μg) and IL-17A (0.5 μg) every third day for a total of nine days and eWAT RNA isolated. Il33 transcript levels were measured by quantitative real-time PCR and normalized to Tbp (n ≥ 5). Representative flow cytometry plots (c) and Il33 expression from iWAT stromal cells (d) after WT mice were injected with saline or TNF (1 μg) and IL-17A (0.5 μg) every third day for a total of nine days. Il33 normalized with Tbp (n ≥ 3, pooled). Small horizontal lines indicate the mean. ** P < 0.01; **** P < 0.0001 (One-way ANOVA in a,d; Student’s t test in b). Data are pooled across two experiments run in triplicates (a; mean ± s.e.m. in a). Data are representative of two experiments (b-d; mean ± s.e.m. in b,d).
Supplementary Figure 4 Decreased numbers, and not gene expression, probably contribute to lower IL-33 protein.
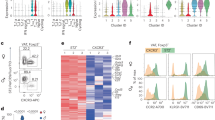
(a) Quantification of numbers (top) and frequencies (bottom) of CD31+, PDGFRα+Pdpn–, Pdpnhi, and Pdpnlo eWAT stromal cells from 23 wk old WT, Tcrd–/–, Vg4/6–/–, and Il17a–/– male mice (n ≥ 3 mice per genotype). (b) Quantitative real-time PCR for Il33 expression normalized with Tbp from sorted Pdpnhi, PDGFRα+, CD31+, and CD45+ cells from WT, Tcrd–/–, Vg4/6–/–, and Il17a–/– mice (n ≥ 3 mice per genotype). Each symbol represents an individual mouse; small horizontal lines indicate the mean. NS, not significant (P > 0.05); * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. (One-way ANOVA in a-b). Data are representative of two experiments (a-b; mean ± s.e.m. in a-b).
Supplementary Figure 5 γδ T cells promote temperature regulation and IL-33 homeostasis in BAT and iWAT.
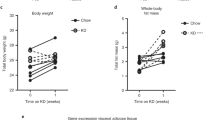
(a) IL-33 protein was quantified from cell lysates of eWAT, iWAT, and BAT from WT, Tcrd–/– and Vg4/6–/– mice using ELISA (left). Quantitative real-time PCR for Il33 expression normalized with Tbp (right) from iWAT and BAT of WT, Tcrd–/– and Vg4/6–/– mice (n ≥ 4). (b) Representative gross anatomy of iWAT from 22 wk old WT, Tcrd–/– and Vg4/6–/– mice after 6 h at 4 °C. (c) Energy expenditure measured from WT and Tcrd–/– mice injected with sterile saline at time 0 h and subsequently injected with selective β3-adrenergic receptor, CL-316 243, (1mg/kg) at 3 h (n = 5 per genotype). Small horizontal lines indicate the mean. NS, not significant; * P < 0.05; ** P < 0.01; *** P < 0.001. (One-way ANOVA in a; Metabolic variable adjusted for differences in body composition by ANCOVA in c). Data are representative of two experiments (a,b; mean ± s.e.m. in a) or one experiment (c; mean ± s.e.m. in c).
Supplementary Figure 6 IL-17A promotes thermogenic responses in BAT and iWAT.
(a) Frequency (left) and numbers (right) of γδ T cells at 0, 8, and 24 h at 4 °C in BAT and iWAT (n ≥ 3 mice per condition). (b) Quantitative real-time PCR of Ppargc1a, Dio2, and Cox7a1 normalized to Tbp in BAT between WT and Il17a–/– mice (n ≥ 3). (c) Quantitative real-time PCR of Ppargc1a and Dio2 normalized to Tbp in iWAT between WT and Il17a–/– mice (n ≥ 3). (d) Mice were gradually shifted from 30 °C to 4 °C at a continuous rate and body temperature measured between WT and Il17a–/– male mice (n = 5 mice per genotype). (e) Body temperature (top) and RER (bottom) measured for 72 h at thermoneutrality after acclimation between WT and Il17a–/– male mice (n = 5 per genotype). Each symbol represents an individual mouse; small horizontal lines indicate the mean. NS, not significant (P > 0.05); * P < 0.05; ** P < 0.01; *** P < 0.001. (Student’s t test in b-c; One-way ANOVA in a; Metabolic variable adjusted for differences in body composition by ANCOVA in d-e). Data are representative of two experiments (a-c; mean ± s.e.m. in a-c).
Supplementary Figure 7 Gene expression analysis of BAT and iWAT.
Quantitative real-time PCR of Th, Adrb3, Lipe (Hsl), and Pnpla2 (Atgl) in brown (a) and inguinal (b) adipose tissue obtained from WT, Tcrd–/–, Vg4/6–/– and Il17a–/– mice at room temperature (25 °C) and after 6 h cold at 4 °C. Genes normalized to Tbp (n ≥ 4 mice per condition). Each symbol represents an individual mouse; small horizontal lines indicate the mean. NS, not significant (P > 0.05); * P < 0.05; ** P < 0.01. (One-way ANOVA in a,b). Data are representative of two experiments (a,b; mean ± s.e.m. in a,b).
Supplementary Figure 8 γδ T cells directly and indirectly influence adaptive thermogenesis.
(a) Differentiated brown adipocytes were stimulated with indicated amounts of TNFlo (0.1ng/mL), TNFhi (1ng/mL), IL-17Alo (0.1ng/mL), IL-17Ahi (1ng/mL), for 18 h and Ucp1, Dio2, Cidea, and Il33 transcript levels were measured by quantitative real-time PCR and normalized with Tbp. (b) Differentiated brown adipocytes were stimulated with either IL-33lo (10ng/mL), IL-33hi (100ng/mL), and analyzed as in a. (c) Representative flow cytometry plots (left) of iWAT stromal cells after WT mice were injected with saline (top row) or TNF (1 μg) and IL-17A (0.5 μg) every third day for a total of nine days. Pdpn+PDGFRα– and PDGFRα+ iWAT stromal cells were sorted and gene expression of Ucp1, Ppargc1a, and Dio2 measured by quantitative real-time PCR and normalized with Tbp (n ≥ 3). Frequency (top) and numbers (bottom) of eosinophils, ILC2s, iNKT, and Treg cells from WT, Tcrd–/– and Vg4/6–/– brown (d) and inguinal (e) adipose tissue from 22 wk male mice (n ≥ 4 mice per group). Each symbol represents an individual replicate or mouse. Data are representative of two experiments (a-e; mean ± s.e.m. in a-e). NS, not significant (P > 0.05); * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. (One-way ANOVA in a-e).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8
Rights and permissions
About this article
Cite this article
Kohlgruber, A.C., Gal-Oz, S.T., LaMarche, N.M. et al. γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat Immunol 19, 464–474 (2018). https://doi.org/10.1038/s41590-018-0094-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0094-2
This article is cited by
-
Cytokine production by bovine adipose tissue stromal vascular fraction cells upon Neospora caninum stimulation
Scientific Reports (2024)
-
The chemerin-CMKLR1 axis in keratinocytes impairs innate host defense against cutaneous Staphylococcus aureus infection
Cellular & Molecular Immunology (2024)
-
Interplay of IL-33 and IL-35 Modulates Th2/Th17 Responses in Cigarette Smoke Exposure HDM-Induced Asthma
Inflammation (2024)
-
IL-17 signalling is critical for controlling subcutaneous adipose tissue dynamics and parasite burden during chronic murine Trypanosoma brucei infection
Nature Communications (2023)
-
γδ T cells control murine skin inflammation and subcutaneous adipose wasting during chronic Trypanosoma brucei infection
Nature Communications (2023)