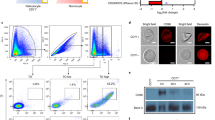
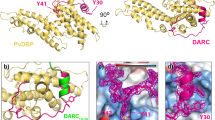
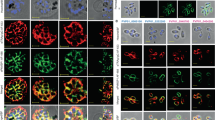
Abstract
Plasmodium vivax is the most widely distributed malaria parasite that infects humans1. P. vivax invades reticulocytes exclusively, and successful entry depends on specific interactions between the P. vivax reticulocyte-binding protein 2b (PvRBP2b) and transferrin receptor 1 (TfR1)2. TfR1-deficient erythroid cells are refractory to invasion by P. vivax, and anti-PvRBP2b monoclonal antibodies inhibit reticulocyte binding and block P. vivax invasion in field isolates2. Here we report a high-resolution cryo-electron microscopy structure of a ternary complex of PvRBP2b bound to human TfR1 and transferrin, at 3.7 Å resolution. Mutational analyses show that PvRBP2b residues involved in complex formation are conserved; this suggests that antigens could be designed that act across P. vivax strains. Functional analyses of TfR1 highlight how P. vivax hijacks TfR1, an essential housekeeping protein, by binding to sites that govern host specificity, without affecting its cellular function of transporting iron. Crystal and solution structures of PvRBP2b in complex with antibody fragments characterize the inhibitory epitopes. Our results establish a structural framework for understanding how P. vivax reticulocyte-binding protein engages its receptor and the molecular mechanism of inhibitory monoclonal antibodies, providing important information for the design of novel vaccine candidates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mueller, I. et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect. Dis. 9, 555–566 (2009).
Gruszczyk, J. et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science 359, 48–55 (2018).
Tham, W.-H. et al. Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proc. Natl Acad. Sci. USA 107, 17327–17332 (2010).
Horuk, R. et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science 261, 1182–1184 (1993).
Miller, L. H., Mason, S. J., Dvorak, J. A., McGinniss, M. H. & Rothman, I. K. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189, 561–563 (1975).
Adams, J. H. et al. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell 63, 141–153 (1990).
Cowman, A. F., Tonkin, C. J., Tham, W.-H. & Duraisingh, M. T. The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe 22, 232–245 (2017).
Maier, A. G. et al. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 9, 87–92 (2003).
Crosnier, C. et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 480, 534–537 (2011).
WHO. World Malaria Report 2016. WHO http://www.who.int/malaria/publications/world-malaria-report-2016/report/en/. (2017)
Hegner, R. Relative frequency of ring-stage plasmodia in reticulocytes and mature erythrocytes in man and monkey. Am. J. Epidemiol. 27, 690–718 (1938).
Carlton, J. M. et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455, 757–763 (2008).
Galinski, M. R., Medina, C. C., Ingravallo, P. & Barnwell, J. W. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell 69, 1213–1226 (1992).
Gruszczyk, J. et al. Structurally conserved erythrocyte-binding domain in Plasmodium provides a versatile scaffold for alternate receptor engagement. Proc. Natl Acad. Sci. USA 113, E191–E200 (2016).
Lawrence, C. M. et al. Crystal structure of the ectodomain of human transferrin receptor. Science 286, 779–782 (1999).
Lebron, J. A. et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell 93, 111–123 (1998).
Abraham, J., Corbett, K. D., Farzan, M., Choe, H. & Harrison, S. C. Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat. Struct. Mol. Biol. 17, 438–444 (2010).
Radoshitzky, S. R. et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446, 92–96 (2007).
Cheng, Y., Zak, O., Aisen, P., Harrison, S. C. & Walz, T. Structure of the human transferrin receptor-transferrin complex. Cell 116, 565–576 (2004).
Pearson, R. D. et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat. Genet. 48, 959–964 (2016).
Demogines, A., Abraham, J., Choe, H., Farzan, M. & Sawyer, S. L. Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol. 11, e1001571 (2013).
Radoshitzky, S. R. et al. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc. Natl Acad. Sci. USA 105, 2664–2669 (2008).
Mikolajczak, S. A. et al. Plasmodium vivax liver stage development and hypnozoite persistence in human liver-chimeric mice. Cell Host Microbe 17, 526–535 (2015).
Neafsey, D. E. et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat. Genet. 44, 1046–1050 (2012).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007).
Yang, N., Zhang, H., Wang, M., Hao, Q. & Sun, H. Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe. Sci. Rep. 2, 999 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Harding, S. E., Rowe, A, J., Horton, J. C. (eds.) Analytical Ultracentrifugation in Biochemistry and Polymer Science (Royal Society of Chemistry, Cambridge, 1992).
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Kantardjieff, K. A. & Rupp, B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein–nucleic acid complex crystals. Protein Sci. 12, 1865–1871 (2003).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Painter, J. & Merritt, E. A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Cryst. 39, 109–111 (2006).
Birmanns, S., Rusu, M. & Wriggers, W. Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J. Struct. Biol. 173, 428–435 (2011).
Acknowledgements
We thank J. Newman from CSIRO Collaborative Crystallization Centre for assistance with setting up the crystallization screens, the Walter and Eliza Hall Institute’s Monoclonal Antibody Facility for production of antibodies, and the MX and SAXS beamline staff at the Australian Synchrotron for their assistance during data collection. This work was supported in part by the Australian Research Council Future Fellowship to W.-H.T. and M.D.W.G., a Speedy Innovation Grant to W.-H.T. and a National Health and Medical Research Council fellowship (1105754) to J.M.M. W.-H.T. is a Howard Hughes Medical Institute–Wellcome Trust International Research Scholar (208693/Z/17/Z). R.D.P. is funded by Wellcome Trust (090770). The authors acknowledge support from the Drakensberg Trust, the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
Reviewer information
Nature thanks S. Scheres and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
J.G., L.-J.C. and S.M. established experimental procedures. J.G. and W.W. optimized the samples for cryo-EM. R.K.H., C.H. and Z.Y. performed specimen preparation for cryo-EM, collected and processed the data and generated the maps. J.G. carried out model building, refinement and analysed the structure of PvRBP2b–TfR1–Tf complex. J.G., L.-J.C. and S.M. expressed and purified all mutants and performed the structure–function analyses. L.-J.C. and J.G. purified the Fab fragments. J.G. crystallized, collected data and solved the structures of Fab complexes. J.M.M. and J.G. collected and analysed SAXS data. Y.-F.M. and M.D.W.G. collected and analysed AUC data. R.D.P. performed the sequence diversity analyses. W.-H.T., J.G. and A.F.C. designed the project. W.-H.T., J.G. and Z.Y. analysed the data, and W.-H.T. and J.G. principally wrote the manuscript which was finalized with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Details of cryo-EM data collection and analysis.
a, Coomassie-stained reducing SDS–PAGE gel of proteins used for the sample preparation. M, molecular weight marker. For gel source data, see Supplementary Fig. 1. b, Representative micrograph of the sample after drift correction and dose weighing. c, Representative 2D class averages. The 2D class averages exhibit different projections corresponding to each orientation. d, 3D classification effectively separated two groups of populations. Left, TfR1–Tf complex with one bound PvRBP2b molecule. Right, TfR1–Tf complex with two bound PvRBP2b molecules. Blue, TfR1–Tf; pink, PvRBP2b. Scale bar, 10 Å. e, Local resolution estimation diagram of the final refined maps. Left, the one-ligand complex; right: the two-ligand complex. Resolution keys are labelled from 3.5 to 6.0 Å. f, Resolution estimation of the cryo-EM map. Fourier shell correlation (FSC) plot showing resolutions at 0.143 FSC (dashed line) for both complexes, determined by the gold-standard method. g, Representative cryo-EM density for different parts of the C-terminal domain of PvRBP2b. Selected residues are also indicated. h, FSC curves of the final refined model versus the final cryo-EM map (full dataset, blue), of the outcome of model refinement with a half map versus the same map (red), and of the outcome of model refinement with a half map versus the other half map (green). In a–c, the experiment was performed once.
Extended Data Fig. 2 Detailed structural analyses of PvRBP2b and sedimentation velocity analysis of the formation of the ternary PvRBP2b(161–969)–TfR1–Tf complex.
a, Superposition of the cryo-EM structure of PvRBP2b with the previously reported crystal structure of the N-terminal domain (PDB ID: 5W53). b, Two orthogonal views of PvRBP2b shown in surface representation and coloured according to the potential on the surface of the molecule. Electrostatic surface potential was calculated using the program APBS in Chimera with the nonlinear Poisson–Boltzmann equation and contoured at ± 5 kT e−1. Negatively and positively charged surface areas are coloured red and blue, respectively. c, Continuous sedimentation coefficient (c(s)) distributions for the TfR1–Tf complex (2.5 μM complex) in the presence of increasing concentrations of PvRBP2b(161–969) (0.25–16 μM). The configuration and surface charge properties of the ternary complex suggest that this particle may display non-ideal sedimentation, possibly contributing to the observed shift in sedimentation coefficient. Thus, the integrated absorbance signal in the fast-sedimenting peak of the distributions was analysed as a function of PvRBP2b(161–969) concentration. d, Here the TfR1–Tf complex was assumed to sediment as a single, stable species. These data are consistent with the binding of two molecules of PvRBP2b(161–969) to the TfR1–Tf complex with macroscopic dissociation constants in the mid-nanomolar to low-micromolar range. e, Residuals for the best fit of the raw radial absorbance sedimentation velocity data to a c(s) distribution model for 2.5 μM of TfR1–Tf in the presence of (top to bottom): 0.25 μM, 0.5 μM, 1.0 μM, 2.0 μM, 4.0 μM, 8.0 μM and 16.0 μM PvRBP2b(161–969).
Extended Data Fig. 3 Structure of the one-ligand complex and conformational changes induced in TfR1 upon PvRBP2b binding.
a, Overall view of the one-ligand complex. Proteins are shown in ribbon representation. The molecule of TfR1 interacting with PvRBP2b is coloured according to domain organization. Protease-like domain, red; apical domain, green; helical domain, yellow. The other molecule of TfR1 that remains unliganded is coloured in wheat. Tf, cyan; Fe3+, red spheres. PvRBP2b N-terminal domain, violet; PvRBP2b C-terminal domain, blue. b, Superimposition of the unliganded TfR1 molecule from the one-ligand complex on the ligand-bound TfR1 from the two-ligand complex. Colour-coded as in a. The movement of the apical domain is indicated schematically by a black arrow. For clarity, both Tf molecules have been omitted. c, Closer view of the superimposition shown in b. The residues that are most affected by PvRBP2b binding are shown as sticks and labelled. d, The superposition of TfR1 in complex with Tf and PvRBP2b (coloured according to domain), and TfR1 in the complex with Tf only (grey, PDB ID: 3S9L). The r.m.s.d. between the 1,090 aligned Cα atoms is 0.81 Å. The movement of the apical domain is represented schematically with black arrows. e, Superposition of Tf in the cryo-EM structure of PvRBP2b–TfR1–Tf complex (in cyan) with the crystal structure of holo-Tf in Tf–TfR1 complex (left, wheat, PDB ID: 3S9L) or with the crystal structure of holo-Tf (middle, green, PDB ID: 3V83) or with the structure of apo-Tf (right, orange, PDB ID: 2HAV). Fe3+, red spheres. R.m.s.d. values are indicated above each superimposition. f, Two orthogonal views of the superposition between two subclasses for the two-ligand complex. Molecules are shown in cartoon representation and coloured in violet and cyan for subclasses 1 and 2, respectively. The most important difference between the two subclasses is the movement of the N-terminal domain of PvRBP2b, indicated by the black arrows. g, Close view of the PvRBP2b–Tf interaction site. The N2 subdomain of Tf in the cryo-EM structure (cyan) is superimposed in the crystal structures of holo-Tf in closed conformation (wheat), holo-Tf in partially open conformation (green) and apo-Tf in open conformation (orange). The movement of the N1 subdomain is highlighted schematically with black arrows.
Extended Data Fig. 4 Site-directed mutagenesis of PvRBP2b and TfR1.
a, SDS–PAGE of purified PvRBP2b mutant recombinant proteins. Two micrograms of each protein were loaded onto a 4–12% NuPAGE gradient gel under reducing conditions and stained with Coomassie blue. b, Circular dichroism spectra of recombinant PvRBP2b mutants. c, Analytical SEC (left to right panel) of complex formation between TfR1–Tf and PvRBP2b with mutant residues Y538, Y542, K600 or Y604. SDS–PAGE of fractions from the SEC analyses. d, SDS–PAGE gel of purified TfR1 mutant recombinant proteins. Two micrograms of each protein were loaded onto a 4–12% NuPAGE gradient gel under reducing conditions and stained with Coomassie blue. e, Analytical SEC of TfR1 mutants in complex with Tf. f, Circular dichroism spectra of recombinant TfR1 mutants. All experiments were performed once. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 Most TfR1 residues that interact with PvRBP2b are specific to human TfR1.
a, Schematic representation of TfR1 sequence coloured according to domain. Apical domain, green; protease-like domain, red; helical domain, yellow. Domain boundaries are indicated above the schematic. The most important residues forming either hydrogen bonds or salt bridges with PvRBP2b are indicated below with black arrows. b, Comparison between Machupo virus GP1 and PvRBP2b binding sites. Human TfR1 is shown in the centre in surface representation and coloured according to the domain organization. The residues that interact with PvRBP2b are shown in violet and those that interact with Machupo virus GP1 are dark blue. Residues overlapping the two sites are shown in magenta. The surrounding panels show fragments of the alignment between TfR1 sequences from different species (human, mouse, rat, hamster, cat and dog). Residues are coloured according to their chemical properties (acidic, red; basic, blue; polar, magenta; cysteine, yellow; hydrophobic, grey scale with intensity proportional to hydrophobicity). The fragments of the alignment corresponding to the apical and protease-like domains of TfR1 are additionally shaded in green and red, respectively. The residues interacting with PvRBP2b are indicated with stars above the alignment, which are coloured according to the type of interaction (hydrogen bonds, green; salt bridges, red; others, empty stars). The Machupo virus GP1-interaction site is indicated with a dark blue line. The position of the G217 deletion and the G142S polymorphism are indicated with black arrows above the alignment. c, Analytical SEC results showing PvRBP2b(161–1454) interaction with mouse Tf and TfR1. For gel source data, see Supplementary Fig. 1. Top, elution profiles for the separate components and for the combined components. Bottom, SDS–PAGE of SEC fractions. Left, human Tf and human TfR1; middle left, mouse Tf and human TfR1; middle right, human Tf and mouse TfR1; right, mouse Tf and mouse TfR1. The same representative SDS–PAGE gel is used for PvRBP2b protein SEC analyses. All analytical SEC experiments were performed once.
Extended Data Fig. 6 Reticulocyte-binding and SAXS data analyses for the N-terminal domain of PvRBP2b in complexes with four different inhibitory Fab fragments.
a, PvRBP2b(161–1454) binding in the presence or absence of anti-PvRBP2b monoclonal antibodies (3E9, 4F7, 6H1, 8G7 and 10B12) or anti-PfRh4 monoclonal antibodies (10C9), analysed by flow cytometry. Dot plots of PvRBP2b(161–1454) binding (y axis) to reticulocytes stained with thiazole orange (TO, x axis). This experiment was repeated independently four times with similar results. b, Arbitrarily offset scattering intensity profiles for the PvRBP2b–Fab complexes. The background-subtracted SAXS data are shown as black open circles representing natural logarithm of mean intensity lnI(q) as a function of momentum transfer q in Å−1. a.u., arbitrary unit. The theoretical scattering profiles (solid lines: PvRBP2b–3E9, blue; PvRBP2b–4F7, red; PvRBP2b–6H1, green; PvRBP2b–10B12, yellow) calculated from the crystal structures were fitted to the experimental scattering data using CRYSOL. c, Guinier plots for qRg ≤ 1.3 (qRg is the product of momentum transfer (q) and the radius of gyration (Rg)), showing that neither high-molecular-mass aggregates nor inter-particle interference contribute measurably to scattering and the data are of high quality (colours as in b). d, Pair-wise inter-atomic distance distribution function, P(r) (colours as in b). e, Two orthogonal views of ab initio bead models represented as grey spheres, superimposed with the crystal structure of the corresponding antibody complexes. For the PvRBP2b–10B12 complex, COLLAGE was used to rigid-body fit two crystal structures, the N-terminal domain of PvRBP2b (PDB ID: 5W53) and the structure of the 10B12 Fab fragment alone. f, Table summarizing SAXS data collection and analysis. The radius of gyration and initial scattering intensity I(0) were approximated using the Guinier equation with PRIMUS. The radius of gyration and maximum particle dimension Dmax were calculated from the P(r) analysis using program GNOM. The normalized spatial discrepancy parameter (NSD) indicates the similarity between 20 independently generated ab initio models. The χ parameter provides the assessment of the fit of the average model to the experimental data calculated using program CRYSOL. The structural parameters and their associated errors are those derived by least-squares curve-fitting analysis of the scattering data as formulated within the listed software packages.
Supplementary information
Supplementary Figure 1
This figure shows the original scans of all the representative gels. The lined boxes indicate parts used for the figures.
Rights and permissions
About this article
Cite this article
Gruszczyk, J., Huang, R.K., Chan, LJ. et al. Cryo-EM structure of an essential Plasmodium vivax invasion complex. Nature 559, 135–139 (2018). https://doi.org/10.1038/s41586-018-0249-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0249-1
This article is cited by
-
Course of Plasmodium infection studied using 2D-COS on human erythrocytes
Malaria Journal (2023)
-
Genetic diversity of Plasmodium vivax reticulocyte binding protein 2b in global parasite populations
Parasites & Vectors (2022)
-
The mutual crosstalk between iron and erythropoiesis
International Journal of Hematology (2022)
-
Naturally acquired blocking human monoclonal antibodies to Plasmodium vivax reticulocyte binding protein 2b
Nature Communications (2021)
-
The genome of the zoonotic malaria parasite Plasmodium simium reveals adaptations to host switching
BMC Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.