Abstract
Mono-allelic germline pathogenic variants in the Partner And Localizer of BRCA2 (PALB2) gene predispose to a high-risk of breast cancer development, consistent with the role of PALB2 in homologous recombination (HR) DNA repair. Here, we sought to define the repertoire of somatic genetic alterations in PALB2-associated breast cancers (BCs), and whether PALB2-associated BCs display bi-allelic inactivation of PALB2 and/or genomic features of HR-deficiency (HRD). Twenty-four breast cancer patients with pathogenic PALB2 germline mutations were analyzed by whole-exome sequencing (WES, n = 16) or targeted capture massively parallel sequencing (410 cancer genes, n = 8). Somatic genetic alterations, loss of heterozygosity (LOH) of the PALB2 wild-type allele, large-scale state transitions (LSTs) and mutational signatures were defined. PALB2-associated BCs were found to be heterogeneous at the genetic level, with PIK3CA (29%), PALB2 (21%), TP53 (21%), and NOTCH3 (17%) being the genes most frequently affected by somatic mutations. Bi-allelic PALB2 inactivation was found in 16 of the 24 cases (67%), either through LOH (n = 11) or second somatic mutations (n = 5) of the wild-type allele. High LST scores were found in all 12 PALB2-associated BCs with bi-allelic PALB2 inactivation sequenced by WES, of which eight displayed the HRD-related mutational signature 3. In addition, bi-allelic inactivation of PALB2 was significantly associated with high LST scores. Our findings suggest that the identification of bi-allelic PALB2 inactivation in PALB2-associated BCs is required for the personalization of HR-directed therapies, such as platinum salts and/or PARP inhibitors, as the vast majority of PALB2-associated BCs without PALB2 bi-allelic inactivation lack genomic features of HRD.
Similar content being viewed by others
Introduction
The Partner And Localizer of BRCA2 (PALB2) is a key protein that interacts with BRCA1 and BRCA2 and plays pivotal roles in homologous recombination (HR) DNA repair.1 Bi-allelic PALB2 germline mutations (i.e., affecting both parental alleles of PALB2) cause Fanconi anemia,2 whereas mono-allelic PALB2 germline mutations result in increased risk of breast, pancreatic and ovarian cancer.3,4,5 The frequency of PALB2 germline mutations in familial breast cancer ranges from 0.6% to 2.7%,4 and the average cumulative breast cancer risk in PALB2 germline mutation carriers by the age of 70 years is ~35%,4 similar to that conferred by BRCA2 germline mutations.6 Akin to sporadic and BRCA2 breast cancers, PALB2-associated breast cancers are heterogeneous in terms of their clinicopathologic features, being predominantly estrogen receptor (ER)-positive.4 As compared to non-PALB2 mutation carriers, patients with PALB2 germline mutations have been reported to display a shorter 10-year survival.7 Consistent with the role of PALB2 in HR DNA repair, PALB2-deficient cells have been shown to be sensitive to platinum-based chemotherapy and poly (ADP-ribose) polymerase (PARP) inhibitors;8,9 hence, therapies targeting HR deficiency (HRD) may benefit breast cancer patients with PALB2 germline mutations.10
Although PALB2 constitutes a tumor suppressor gene, there is controversy as to whether it follows the Knudson two-hit model.11,12 A recent study revealed that ten of 15 PALB2-associated breast cancers harbored bi-allelic PALB2 inactivation through somatic loss of heterozygosity (LOH) of the PALB2 wild-type allele (n = 6) or somatic PALB2 mutations (n = 4).12 PALB2 promoter hypermethylation has been reported in tumors from sporadic and BRCA1/2 mutation-negative familial breast and ovarian cancers,13 however, it appears to be vanishingly rare in tumors from PALB2 germline mutation carriers.12,14 Germane to the understanding of the biology of PALB2-associated breast cancers and to the identification of optimal therapeutic approaches for patients with PALB2 germline mutations is to ascertain the mechanisms that contribute to bi-allelic PALB2 inactivation, and to define whether PALB2-associated breast cancers without bi-allelic inactivation lack genomic features consistent with HRD (e.g., large-scale state transitions (LSTs) and mutational signatures). Importantly, Lee et al.,12 based on a targeted capture sequencing analysis of 487 genes, reported that, with one exception, PALB2-associated breast cancers that retained the PALB2 wild-type allele displayed HRD scores consistent with those of tumors harboring PALB2 bi-allelic inactivation.
Here we sought to characterize the repertoire of somatic genetic alterations of breast cancers from pathogenic PALB2 germline mutation carriers using a combination of whole-exome and targeted massively parallel sequencing to define whether bi-allelic PALB2 inactivation is present in these tumors. Based on whole-exome sequencing (WES) results, we employed validated approaches to determine whether the genomic hallmark features of HRD15 are present in PALB2-associated breast cancers irrespective of the presence of a second hit affecting PALB2. Moreover, as an exploratory, hypothesis-generating analysis, we compared the genomic landscape of breast cancers from pathogenic PALB2 germline mutation carriers to that of breast cancers arising in BRCA1 or BRCA2 germline mutation carriers, and non-BRCA1/2/PALB2-associated breast cancers.
Results
Clinicopathologic features of PALB2-associated breast cancers
Twenty-four invasive breast cancers from carriers of fourteen distinct pathogenic PALB2 germline mutations4,9,16,17,18 were included in this study. Fourteen cases were subjected to WES and WES sequencing data from two cases were retrieved from TCGA19 (n = 16; median depth of tumor 112 × (range 33 × −289 ×) and normal 129 × (range 46 × −247 ×) samples). In addition, 8 cases were analyzed by targeted capture massively parallel sequencing using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) sequencing assay20 (median depth of tumor 232 × (range 73 × −904 ×), and normal 545 × (range 172 × −1452 ×) samples; Table 1, Supplementary Table 1). All samples included in this study were derived from formalin-fixed paraffin-embedded (FFPE) material. Sample quality was evaluated and was found to be appropriate for the analyses conducted (Supplementary Table 2). All but one PALB2 germline mutations were bona fide loss-of-function (frameshift or truncating) mutations; one case carried a missense substitution (L35P), which we have previously demonstrated to be pathogenic.9
The median age at breast cancer diagnosis was 49 years (range 29–82 years), and the median tumor size was 1.6 cm (range 0.15–6 cm; Table 1). All PALB2-associated breast cancers were invasive ductal carcinomas of no special type, and one, 12, and eleven cases were of histologic grades 1, 2, and 3, respectively (Table 1). Eighteen cases (75%) were ER-positive/HER2-negative (ER+/HER2−), five (21%) were ER-negative/HER2-negative (ER−/HER2−), and one (4%) was ER-positive/HER2-positive (ER+/HER2+; Table 1). Whilst the distribution of PALB2-associated breast cancers into ER/HER2 clinical subgroups was found to be comparable to that of sporadic and BRCA2 breast cancers,19,21 it differed from that of BRCA1 breast cancers, which are preferentially of triple-negative phenotype (approximately 70%–85%).21–23
Repertoire of somatic genetic alterations in PALB2-associated breast cancers
Somatic mutation analysis of the WES data of 16 PALB2-associated breast cancers revealed a median of 113.5 (range 59–269) somatic mutations per case, of which 82.5 (range 37–195) were non-synonymous. The eight PALB2-associated breast cancers analyzed by MSK-IMPACT displayed a median of 3 (range 0–5) somatic mutations per case, of which 2 (range 0–4) were non-synonymous (Table 1, Supplementary Table 3). Selected somatic mutations (n = 17) were validated by Sanger sequencing (Supplementary Fig. 1).
Of the 410 cancer genes included in MSK-IMPACT, recurrently mutated genes found in the 24 PALB2-associated breast cancers included PIK3CA (n = 7, 29%), PALB2 (n = 5, 21%), TP53 (n = 5, 21%), NOTCH3 (n = 4, 17%), KMT2A (n = 3, 17%) and ARID1A (n = 3, 17%; Fig. 1, Supplementary Table 3). Six PIK3CA missense mutations affected hotspot residues, including H1047R (n = 3), E545K (n = 1), N345K (n = 1) and C420R (n = 1), and four were predicted to be clonal (Supplementary Fig. 2, Supplementary Table 3). All but one somatic TP53 mutation, all coupled with loss of heterozygosity (LOH) of the TP53 wild-type allele, were predicted to be clonal (Supplementary Fig. 2, Supplementary Table 3). Additional recurrently mutated genes detected in the 16 PALB2-associated breast cancers profiled by WES included CTNNA2, TMPRSS13, KRTAP4–11, LAMA5, KALRN, and COLL22A1 (all, n = 3; Supplementary Fig. 3, Supplementary Table 3).
Non-synonymous somatic mutations in PALB2-associated breast cancers. Heatmap depicting the somatic genetic alterations identified in the 24 PALB2-associated breast cancers analyzed by whole-exome (n = 16) or targeted MSK-IMPACT (n = 8) massively parallel sequencing. Somatic mutations affecting the 410 cancer genes present in MSK-IMPACT, in decreasing overall mutational frequency observed in PALB2-associated breast cancers are plotted. Cases are shown in columns, and genes in rows. Estrogen receptor (ER) and HER2 status, PALB2 germline mutation type, presence of a second somatic PALB2 mutation or loss of heterozygosity (LOH) of the PALB2 wild-type allele, large-scale state transition (LST) score, dominant mutational signature and sequencing platform are indicated in the phenobar (top), color-coded according to the legend. Note that mutational signatures and LST scores could not be assessed in tumors subjected to MSK-IMPACT sequencing due to the limited number of mutations present. Clonal somatic PALB2 mutations or clonal LOH of the PALB2 wild-type allele are indicated by yellow boxes. Somatic mutations are color-coded according to the legend, and LOH of the wild-type allele of mutated genes other than PALB2 is represented by a diagonal bar. Indel small insertion/deletion; LOH loss of heterozygosity, LST large-scale state transition, N/A not assessable, SNV single nucleotide variant, WES whole-exome sequencing
Copy number (CN) analysis revealed recurrent gains of 1q, 8q, 16p, 17q, and 20q, and losses of 1p, 4p, 8p, 11p, and 17p in the 24 PALB2-associated breast cancers analyzed (Fig. 2). Although the majority (n = 18) of cases were ER+/HER2−, concurrent 1q gains and 16q losses, the hallmark features of luminal breast cancers,19 were only found in four cases. Loci recurrently amplified included those mapping to 8q21.3 (encompassing the locus of NBN, n = 5) and 8q24.21 (encompassing the locus of MYC, n = 5; Fig. 2).
Repertoire of copy number alterations in PALB2-associated breast cancers. Copy number alterations in the 24 PALB2-associated breast cancers analyzed by whole-exome (n = 16) or targeted MSK-IMPACT (n = 8) massively parallel sequencing. Cases are represented in rows and chromosomes in columns along the x-axis. Immunohistochemical features, PALB2 germline mutation type, presence of a somatic PALB2 mutation or loss of heterozygosity (LOH) of the PALB2 wild-type allele, large-scale state transition (LST) score, mutational signature and sequencing platform are provided in the phenobar (left), color-coded according to the legend. Dark red, amplification; light red, copy number gain; dark blue, homozygous deletion; light blue, copy number loss; white, no change. LOH loss of heterozygosity, LST large-scale state transition; N/A not assessable, SNV single nucleotide variant, WES whole-exome sequencing
Bi-allelic PALB2 inactivation
Bi-allelic PALB2 inactivation was found in 16 of the 24 PALB2-associated breast cancers (67%; Table 1, Fig. 1). In eleven cases, the second hit was in the form of LOH of the PALB2 wild-type allele, whereas in five tumors, it was in the form of an inactivating (i.e., truncating or frameshift) somatic PALB2 mutation. Fifteen of the 16 somatic genetic events leading to bi-allelic inactivation of the PALB2 wild-type allele were predicted to be clonal (Fig. 1, Supplementary Table 3), suggesting that bi-allelic PALB2 inactivation and subsequent complete loss-of-function of PALB2 may constitute an early somatic event in the development of a subset of PALB2-associated breast cancers.
PALB2-associated breast cancers with bi-allelic inactivation display genomic features consistent with HRD
We15 and others24 have demonstrated that bi-allelic inactivation but not mono-allelic alterations of HR-related genes are associated with genomic features consistent with HRD. Hence, we sought to define whether LST scores and dominant mutational signature 3 would be associated with bi-allelic PALB2 inactivation. LST scores and mutational signatures were inferred in the 16 PALB2-associated breast cancers analyzed by WES, of which 13 cases were found to display high LST scores (LSThigh), and nine cases (eight LSThigh and one LSTlow) were found to have a dominant mutational signature 3 associated with HRD (i.e., BRCA1/2 signature; Table 1, Figs. 1 and 3a). No significant association between PALB2 germline mutation types and HRD-related genomic features was observed (P > 0.05; Table 1).
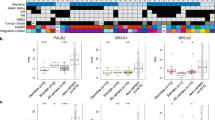
HRD genomic features in breast cancers with and without bi-allelic PALB2 inactivation. a Mutational signatures of all somatic SNVs in the 16 PALB2-associated breast cancers sequenced by whole-exome sequencing (left) as inferred by deconstructSigs41 based on the 30 signatures represented in COSMIC, and a bar plot indicating the proportion of the major mutational signatures identified in each case (right), in decreasing proportion of each signature. The dominant mutational signatures were assigned according to Alexandrov et al.,40 following the consensus of at least two of three approaches (deconstructSigs based on 30 signatures from COSMIC, based on the 12 signatures known to occur in breast cancer, and NMF method42 based on 30 signatures from COSMIC) where signature 1 relates to aging and signature 3 to defective homologous recombination DNA repair, and are shown for cases with bi-allelic PALB2 alterations (top) and mono-allelic PALB2 alterations (bottom). The number of SNVs is shown in parentheses. Sig signature, SNV single nucleotide variant. b Large-scale state transition (LST) scores of the four PALB2-associated breast cancers with mono-allelic PALB2 alterations and the 12 PALB2-associated breast cancers with bi-allelic PALB2 alterations. The median LST scores, and the 75th and 25th percentiles are displayed at the top and bottom of the boxes, respectively. Each dot corresponds to the LST score and the mutational signature of a given case. Dominant mutational signatures are color-coded according to the legend. Comparisons of LST scores between groups were performed using the Mann–Whitney U test. c Average deletion length (nucleotides) in PALB2-associated breast cancers with mono-allelic PALB2 alterations (n = 3) and with bi-allelic PALB2 alterations (n = 11). Only PALB2-associated breast cancers harboring small insertions and deletions were included in the analysis. The median value of deletion length, and the 75th and 25th percentiles are displayed at the top and bottom of the boxes, respectively. Comparisons of deletion lengths between groups were performed using the Mann–Whitney U test. d Number of genes affected by copy number alterations (CNAs) of the four PALB2-associated breast cancers with mono-allelic PALB2 alterations and the 12 PALB2-associated breast cancers with bi-allelic PALB2 alterations. The median value of the number of genes with CNAs, and the 75th and 25th percentiles are displayed at the top and bottom of the boxes, respectively. Comparisons were performed using Fisher’s exact test
Bi-allelic PALB2 inactivation was significantly associated with LSThigh (1/4 vs. 12/12, P = 0.0071, Fisher’s exact test; P = 0.009, Mann–Whitney U test; Fig. 3b), in agreement with the findings that bi-allelic inactivation rather than mono-allelic alterations of HR-related genes is associated with HRD-related genomic features.15,24 All but one (IDC37) cases displaying mutational signature 3 and all but one case (IDC46) displaying high LST scores were found to harbor bi-allelic PALB2 inactivation (Table 1, Fig. 1 and Fig. 3b). Notably, we did not identify pathogenic germline mutations, bi-allelic or mono-allelic somatic mutations or homozygous deletions affecting other HRD-related genes15 in IDC37 or IDC46. Alternative mechanisms of inactivation of the wild-type allele of PALB2, such as complex PALB2 rearrangements or, less likely PALB2 promoter hypermethylation,12 which are not detectable by WES, or other mechanisms that result in HRD may be operative in IDC37 and IDC46. Four of the 12 PALB2-breast cancers with bi-allelic PALB2 inactivation subjected to WES lacked a dominant signature 3, despite displaying high LST scores (Fig. 1). One could hypothesize that these cases could correspond to sporadic breast cancers arising in PALB2 germline mutation carriers, in which the second PALB2 allele was inactivated later in tumor evolution. Two of the 12 PALB2-associated breast cancers analyzed by WES lacked both evidence of bi-allelic PALB2 inactivation and genomic features of HRD (Table 1, Fig. 1). One could posit that these invasive breast cancers may constitute non-PALB2-related cancers arising in the context of a PALB2 germline mutation.
As an exploratory, hypothesis-generating analysis, we compared the genomic profiles of invasive breast cancers developing in the context of pathogenic PALB2 germline mutations with or without loss of the PALB2 wild-type allele. Among the 16 PALB2-associated breast cancers analyzed by WES, the 12 cases with bi-allelic PALB2 inactivation harbored a numerically higher somatic mutation rate (median 139.5, range 63–269) than the four cases without bi-allelic inactivation (median 103, range 59–109; P = 0.09). Moreover, in PALB2-associated breast cancers analyzed by WES and harboring indels (n = 14), the average deletion length was significantly longer in cases with bi-allelic PALB2 inactivation (n = 11) as compared to those with mono-allelic alterations (n = 3; 7 bp vs. 2.3 bp; P = 0.041; Fig. 3c), a feature associated with HRD.25 We further found a significantly higher number of copy number alterations (CNAs) in the tumors with bi-allelic PALB2 inactivation (n = 12) than in those without (n = 4; P = 0.004; Fig. 3d). These results suggest that PALB2-associated breast cancers with bi-allelic inactivation, display higher levels of genetic instability, which may potentially be associated with the early onset of HRD in their development.
Lastly, upon combining the PALB2-associated breast cancers reported by Lee et al.12 with the cases analyzed here, we observed that 67% (26/39) of PALB2-associated breast cancers harbored bi-allelic PALB2 inactivation (Supplementary Table 4). Consistently, bi-allelic PALB2 inactivation was significantly associated with a high LST score, whilst no significant association was observed between bi-allelic PALB2 inactivation and clinicopathologic characteristics (P > 0.05; Supplementary Table 4).
PALB2-associated breast cancers with bi-allelic inactivation display higher mutation burden and HRD-associated features more frequently than sporadic breast cancers
As an exploratory, hypothesis-generating analysis we investigated whether PALB2-associated breast cancers would differ from non-BRCA1/2/PALB2-associated breast cancers from TCGA.19 Given that none of the PALB2-associated breast cancers included here was of ER-/HER2+ phenotype, ER-/HER2+ non-BRCA1/2/PALB2-associated breast cancers from TCGA were excluded, and the remaining 683 ER−/HER2− and ER+ (including ER+/HER2+and ER+/HER2−) breast cancers were employed for the analyses. The 16 PALB2-associated breast cancers analyzed by WES were found to harbor a higher number of somatic mutations (median 113.5, range 59–269) than the 683 ER−/HER2− and ER+non-BRCA1/2/PALB2-associated breast cancers (median 51, range 2–6666; P < 0.002, Mann–Whitney U test), difference that remained significant upon 1:3 bootstrap resampling (P = 0.002, see Methods). Given that the majority of the PALB2-associated breast cancers were ER+/HER2−, we restricted the comparison of mutation burden to the 12 ER+/HER2− PALB2-associated breast cancers sequenced by WES (median of somatic mutations 125, range 63–269) and the 441 ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers (median somatic mutations 42, range 2–6666), and the difference remained significant (P < 0.0001, Mann–Whitney U test; P = 0.0002, bootstrapping-corrected). As expected, the 12 PALB2-associated breast cancers with bi-allelic PALB2 inactivation analyzed by WES (ten ER+/HER2− and two ER−/HER2−) harbored a significantly higher number of somatic mutations (median 139.5, range 63–269) than the 568 ER+/HER2− and ER−/HER2− (median somatic mutations 50, range 2–6666; P = 0.0001, Mann–Whitney U test; P = 0.005, bootstrapping-corrected). It should be noted that no significant differences in the number of somatic mutations were found between the four PALB2-associated breast cancers analyzed by WES lacking bi-allelic PALB2 inactivation (one ER−/HER2− and three ER+; median 103, range 59–109) and the 683 ER−/HER2− and ER+ non-BRCA1/2/PALB2-associated breast cancers (median 51, range 2–6666; P > 0.05, Mann–Whitney U test and bootstrapping-corrected).
A comparison of the frequencies of somatic mutations affecting the 410 cancer genes between PALB2 and non-BRCA1/2/PALB2-associated breast cancers revealed that PALB2, NOTCH3, KMT2A, BRIP1, DNMT3A, FGFR4, GNAQ, and CD79A (all P < 0.05; Fisher’s exact test) were more frequently mutated in the 24 PALB2-associated breast cancers than in the 683 ER−/HER2− and ER+ non-BRCA1/2/PALB2-associated breast cancers (Fig. 4a), however only PALB2 and NOTCH3 remained significantly differently mutated between the two groups after bootstrap resampling (P < 0.01; Supplementary Table 5). No significant differences in the frequency of PIK3CA and TP53 mutations, the two genes most frequently mutated in breast cancer,19 were detected between the 24 PALB2 and the 683 ER−/HER2− and ER+non-BRCA1/2/PALB2-associated breast cancers (Fig. 4a). Upon restriction of the comparison to the 18 ER+ /HER2− PALB2-associated breast cancers and the 441 ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers, PALB2, NOTCH3, ARID1A, DNMT3A, BRIP1, FGFR4, CD79A, and GNAQ (all P < 0.05; Fisher’s exact test) were significantly more frequently mutated in the PALB2-associated breast cancers (Fig. 4b), but similarly only PALB2 and NOTCH3 remained significantly different after bootstrapping resampling (P < 0.01, Fisher’s exact test and bootstrapping-corrected; Supplementary Table 5).
Comparison of PALB2-associated breast cancers and non-BRCA1/2/PALB2-associated breast cancers. a, b Heatmap depicting the most recurrently mutated genes affecting 410 cancer genes identified in PALB2-associated breast cancers and non-BRCA1/2/PALB2-associated breast cancers from TCGA.19 Cases are shown in columns, genes in rows. Multi-Fisher’s exact test comparisons of mutational frequencies of the mutated genes were performed between a the 24 PALB2-associated breast cancers and the 683 ER+/HER2−, ER+/HER2+ and ER−/HER2− non-BRCA1/2/PALB2-associated breast cancers from TCGA, and b the 18 ER+/HER2− PALB2-associated breast cancers and the 441 ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers from TCGA. P-value of each comparison is shown on the right side of the heatmap, with statistically significant P-values in bold. Indel, small insertion/deletion; SNV, single nucleotide variant. c Box and whisker plots showing the large-scale state transition (LST) scores of the PALB2-associated breast cancers with mono-allelic/bi-allelic PALB2 alterations, ER−/HER2− and ER+ non-BRCA1/2/PALB2-associated breast cancers, and ER−/HER2− and ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers where LST scores could be inferred. The median value of LST scores, and the 75th and 25th percentiles are displayed at the top and bottom of the boxes, respectively. Each dot represents the LST score and/or mutational signature of a given case. Mutational signatures are color-coded according to the legend. *of the 601 ER−/HER2− and ER+ non-BRCA1/2/PALB2-associated breast cancers, the 34 cases lacking LST scores but displaying mutational signatures are not shown, three of these cases display signature 3. **of the 491 ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers, the 29 cases lacking LST scores but displaying mutational signatures are not shown, three of these cases display signature 3. P-values of the comparisons of LST scores are shown using Fisher’s exact tests. N/A signatures not assessable, LST large-scale state transition
Differences in the patterns of CNAs were observed between PALB2-associated breast cancers and non-BRCA1/2/PALB2-associated breast cancers. The 24 PALB2-associated breast cancers harbored gains of 16p and losses of 13p and 16q less frequently than the 683 ER−/HER2− and ER+ non-BRCA1/2/PALB2-associated breast cancers (P < 0.05, Fisher’s exact test and bootstrapping-corrected; Supplementary Fig. 4a). When restricting the comparison to the 18 ER+/HER2− PALB2-associated breast cancers and the 441 ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers, the differences were less overt, with more frequent 16p gains and 16q losses in the non-BRCA1/2/PALB2-associated breast cancers (P < 0.05, Fisher’s exact test and bootstrapping-corrected; Supplementary Fig. 4b). Fewer differences were detected in the comparisons between the eight PALB2-associated breast cancers with mono-allelic inactivation (two ER−/HER2− and four ER+) and the 683 ER−/HER2− and ER+non-BRCA1/2/PALB2-associated breast cancers (Supplementary Fig. 4c). No significant difference in the frequency of amplifications and homozygous deletions was observed in any of the comparisons when the bootstrap resampling analysis was performed (P > 0.05; Supplementary Fig. 4d–f).
As part of the exploratory analysis, we investigated whether PALB2-associated breast cancers would differ from non-BRCA1/2/PALB2-associated breast cancers in regard to the frequencies of genomic features indicative of HRD. The 12 PALB2-associated breast cancers with bi-allelic inactivation (two ER−/HER2− and ten ER+/HER2− analyzed by WES) were found to display significantly higher LST scores than the 526 ER−/HER2− and ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers for which LST scores could be determined (P < 0.0001, Mann–Whitney U test; P = 0.0001, bootstrapping-corrected; Fig. 4c). By contrast, the four PALB2-associated breast cancers with mono-allelic inactivation (one ER−/HER2− and three ER+, sequenced by WES) displayed comparable LST scores to the 634 ER−/HER2− and ER+ non-BRCA1/2/PALB2-associated breast cancers for which LST scores could be defined (P > 0.05, Mann–Whitney U test and bootstrapping-corrected; Fig. 4c). Likewise, the proportion of cases displaying a mutational signature 3 was significantly higher in the 12 PALB2-associated breast cancers with bi-allelic inactivation sequenced by WES than in the 491 ER−/HER2− and ER+/HER2− non-BRCA1/2/PALB2-associated breast cancers for which mutational signatures could be inferred (67% vs. 17%; P = 0.0002, Fisher’s exact test; P = 0.02, bootstrapping-corrected; Fig. 4c). These results suggest that PALB2-associated breast cancers with bi-allelic inactivation are more often HR-deficient than non-BRCA1/2/PALB2-associated breast cancers despite displaying a similar prevalence of ER-positive luminal breast cancers, and that PALB2-associated breast cancers without bi-allelic inactivation appear to resemble non-BRCA1/2/PALB2-associated breast cancers.
PALB2-associated breast cancers with bi-allelic inactivation display similarities with BRCA1-associated and BRCA2-associated breast cancers with bi-allelic inactivation of BRCA1/2
Finally, we sought to define whether PALB2-associated breast cancers with bi-allelic PALB2 inactivation would differ from breast cancers arising in BRCA1 and BRCA2 pathogenic germline mutation carriers with bi-allelic inactivation of BRCA1 and BRCA2, respectively. The 12 PALB2-associated breast cancers analyzed by WES were found to harbor a number of somatic mutations (median 139.5, range 63–269) comparable to that of 17 BRCA1-associated breast cancers with bi-allelic inactivation from TCGA (median 143, range 54–1223; P > 0.05, Mann–Whitney U test), and higher than that of the 16 BRCA2-associated breast cancers with bi-allelic inactivation from TCGA (median 74.5, range 38–209; P = 0.006, Mann–Whitney U test). In regards to the repertoire of somatic mutations, PALB2 mutations were significantly more frequent in the 16 PALB2-associated breast cancers with bi-allelic inactivation (n = 5, 31%) than in the 17 BRCA1− (n = 0) and 16 BRCA2-associated (n = 0) breast cancers with bi-allelic inactivation from TCGA (P = 0.02 and P = 0.04, respectively, Mann–Whitney U test; Fig. 5a, b, Supplementary Table 5). In addition, a higher frequency of TP53 mutations was found in the 17 BRCA1-associated breast cancers (n = 15, 88%) than in the 16 PALB2-associated breast cancers with bi-allelic inactivation (n = 3, 19%; P < 0.0001, Fisher’s exact test; Fig. 5a).
Comparison of PALB2-associated breast cancers and BRCA1 and BRCA2 breast cancers. a, b Heatmap depicting the most recurrently mutated genes affecting 410 cancer genes identified in PALB2-associated breast cancers and BRCA1 and BRCA2 breast cancers from TCGA.19 Cases are shown in columns, and genes in rows. Multi-Fisher’s exact test comparisons of mutational frequencies of the recurrently mutated genes were performed between a the 16 PALB2-associated breast cancers with bi-allelic PALB2 alterations and 17 BRCA1 breast cancers bi-allelic BRCA1 alterations, and b the 16 PALB2-associated breast cancers with bi-allelic PALB2 alterations and 16 BRCA2 breast cancers with bi-allelic BRCA2 alterations. P-value of each comparison is shown on the right side of the heatmap, with statistically significant P-values in bold. Indel, small insertion/deletion; SNV, single nucleotide variant. c Boxplots showing the large-scale state transition (LST) scores of the 12 PALB2-associated breast cancers with bi-allelic PALB2 alterations, 17 BRCA1 and 16 BRCA2 breast cancers with bi-allelic BRCA1 and BRCA2 alterations, respectively. The median value of the LST scores, and the 75th and 25th percentiles are displayed at the top and bottom of the boxes, respectively. Each dot corresponds to the LST score and/or mutational signature of one case. Mutational signatures are color-coded according to the legend. P-values of the comparisons of LST scores are shown using Fisher’s exact tests. N/A signatures not assessable, LST large-scale state transition
CN analysis revealed that the 17 BRCA1-associated breast cancers with bi-allelic inactivation had higher frequencies of gains of 3q and 6p and losses of 17q, among other differences (P < 0.05, Fisher’s exact test; Supplementary Fig. 5a), as compared to the 16 PALB2-associated breast cancers with bi-allelic inactivation. In contrast, the CN profiles of the 16 BRCA2 breast cancers with bi-allelic inactivation were more similar to those of the PALB2-associated breast cancers, albeit more frequently harboring losses of 13q and 22q, among other differences (P < 0.05; Supplementary Fig. 5b). No significant difference in the frequency of amplifications and homozygous deletions was found between the 16 PALB2-associated breast cancers and the 17 BRCA1-associated and 16 BRCA2-associated breast cancers with bi-allelic inactivation of the respective wild-type allele (Supplementary Fig. 5c, d).
The LST scores of the 12 breast cancers with bi-allelic PALB2 inactivation analyzed by WES were comparable to those of the breast cancers with bi-allelic BRCA1 inactivation (n = 17) and bi-allelic BRCA2 inactivation from TCGA (n = 16; P > 0.05, Mann–Whitney U test; Fig. 5c), whereas the proportion of the PALB2-associated breast cancers with bi-allelic inactivation displaying signature 3 (67%, 8/12) was not statistically significantly different from that of the BRCA1-associated breast cancers with bi-allelic inactivation (82%, 14/17; P = 0.4, Fisher’s exact test) and BRCA2-associated breast cancers with bi-allelic inactivation from TCGA (63%, 10/16; P = 1, Fisher’s exact test; Fig. 5c). Consistent with these findings, LST score, NtAI score,26 which assesses telomeric allelic imbalance, and the Myriad score,26 which is the unweighted sum of LOH, telomeric allelic imbalance and LSTs, were higher in breast cancers with PALB2 (n = 12) and BRCA1/2 (n = 33) biallelic inactivation, compared to those with PALB2 (n = 4) and BRCA1/2 (n = 8) monoallelic inactivation, respectively (Supplementary Figs. 6a–6c). Moreover, LST score, mutational signature 3, NtAI score and Myriad score detected bi-allelic inactivation of PALB2 and of BRCA1/BRCA2 in PALB2-associated breast cancers and in BRCA1/2-associated breast cancers, respectively, with comparable accuracy (Supplementary Figs. 6d–6g). Taken together, our results suggest that PALB2-associated breast cancers with bi-allelic inactivation are similar to breast cancers with BRCA1 and BRCA2− bi-allelic inactivation in terms of genetic instability and genomic features indicative of HRD.
Discussion
Here we demonstrate that PALB2-associated breast cancers constitute a heterogeneous group of tumors at the genetic level and can be stratified according to the bi-allelic inactivation of the PALB2 wild-type allele. PALB2-associated breast cancers display a high mutation burden and a limited number of genes recurrently affected by pathogenic somatic mutations, including PIK3CA, TP53, NOTCH3, and PALB2 itself. Loss of the PALB2 wild-type allele in PALB2-associated breast cancers occurred in the form of PALB2 pathogenic somatic mutations in five (21%) cases, whereas LOH of the wild-type allele of PALB2 was detected in 11 (46%) cases. Second somatic mutations in BRCA1/2 have been reported as the underlying cause of bi-allelic inactivation in tumors from BRCA1/2 germline mutations carriers.27 It should be noted, however, that somatic mutations resulting in the inactivation of the wild-type allele of BRCA1 or BRCA2 in BRCA1− or BRCA2-associated breast cancers, respectively,15,24,28 appear to be less frequent than somatic PALB2 mutations in the context of PALB2-associated breast cancers. In the study by Maxwell et al.28 bi-allelic BRCA1 inactivation was due to a BRCA1 somatic mutation in only one case (1.1%) out of 93 BRCA1-associated breast and ovarian tumors. Similarly, out of 67 BRCA2-associated tumors with bi-allelic BRCA2 inactivation, in only one case this was due to a BRCA2 somatic mutation (1.5%). In contrast, PALB2 somatic mutations as a mechanism of bi-allelic inactivation were significantly more frequent in the PALB2-associated breast cancers from this series (31%; 5/16; P = 0.00006, Fisher’s exact test).
Consistent with the findings of Lee et al.,12 our study demonstrates that PALB2 follows the Knudson two-hit model, given that in a large proportion of PALB2-associated breast cancers, a second hit in the form of a somatic PALB2 mutation or LOH of the wild-type allele of PALB2 was detected. Contrary to that study,12 in which PALB2-associated breast cancers with either mono-allelic or bi-allelic PALB2 alterations were found to display genomic features of HRD, based on targeted massively parallel sequencing of 487 genes, our WES analysis of 16 PALB2-associated breast cancers revealed that tumors with PALB2 bi-allelic alterations displayed significantly higher LST scores and average deletion lengths than PALB2-associated breast cancers with mono-allelic PALB2 alterations. In addition, only one out of the four PALB2-associated breast cancers with mono-allelic PALB2 alterations displayed a dominant mutational signature 3, whereas eight out of 12 PALB2-associated breast cancers with bi-allelic PALB2 inactivation harbored a dominant mutational signature 3. Our WES findings are consistent with the pan-cancer WES analysis performed by Riaz et al.,15 whereby HR-related genes with bi-allelic inactivation but not those with mono-allelic alterations were found to display genomic features of HRD, and the analyses performed by Polak et al.,24 where bi-allelic, but not mono-allelic, alterations affecting BRCA1, BRCA2, and PALB2 were found to be associated with HRD in breast cancers. Conversely, 8/24 PALB2-associated breast cancers included in this study lacked bi-allelic PALB2 inactivation and 2/16 PALB2-associated breast cancers sequenced by WES lacked both bi-allelic PALB2 inactivation and genomic features of HRD. In this context, one could posit that this subset of PALB2-associated breast cancers may retain competent HR repair of DNA double-strand breaks and would unlikely benefit from HRD-directed therapies. Interestingly, the proportion of PALB2-associated breast cancers displaying mono-allelic PALB2 inactivation was comparable to the one of BRCA1-associated and BRCA2-associated breast cancers from TCGA harboring BRCA1 or BRCA2 mono-allelic inactivation, respectively. Although PALB2 mono-allelic inactivation is not associated with genomic features of HRD, its role in tumorigenesis is yet to be determined.
In agreement with previous studies showing that most breast cancers with HRD features are underpinned by bi-allelic inactivation of HR-related genes,15,29 we identified the genetic basis of HRD in 12 out of 14 (86%) PALB2-associated breast cancers with genomic features of HRD. It should be noted that of the PALB2-associated breast cancers with mono-allelic PALB2 alterations studied here, one displayed a high LST score and another one harbored a dominant mutational signature 3. This observation suggests that other mechanisms of HRD may be operative in these tumors. First, no bi-allelic inactivation of another DNA repair related gene was detected in these cancers. Second, albeit PALB2 gene promoter methylation was reported in two of eight inherited breast cancers and four of 60 sporadic breast cancers,13 this phenomenon appears to be vanishingly rare in PALB2-associated breast cancers with mono-allelic PALB2 alterations.12 Further analyses are warranted to define whether other mechanisms of inactivation of the wild-type allele of PALB2 may play a role in PALB2-associated breast cancers with mono-allelic PALB2 alterations but with genomic features of HRD.
Our study has important limitations. First, as a result of the rarity of PALB2-associated breast cancers, the small sample size may have limited the detection of significant differences in the exploratory analyses comparing PALB2-associated breast cancers with non-BRCA1/2/PALB2-associated breast cancers from TCGA. Importantly, however, these analyses revealed that PALB2-associated breast cancers with bi-allelic inactivation differ from non-BRCA1/2/PALB2-associated breast cancers but are similar to BRCA1 and BRCA2 breast cancers with bi-allelic inactivation. Second, genomic features of HRD were investigated here based on WES; although our approach for the detection of genomic features of HRD is more robust than that based on targeted massively parallel sequencing,12 whole-genome sequencing analysis still remains the optimal approach. Therefore, the proportion of PALB2-associated breast cancers with HRD may be even higher than that reported here and in previous analyses.12
Despite these limitations, our data demonstrate that PALB2-associated breast cancers harbor complex and heterogeneous genomes. Notably, PALB2 bi-allelic inactivation is present in a large proportion of PALB2-associated breast cancers, and the mechanisms leading to this include both LOH of the wild-type allele or pathogenic somatic mutations affecting PALB2. Importantly, the 12 PALB2-associated breast cancers with PALB2 bi-allelic inactivation displayed genomic features consistent with HRD, and shared similarities in terms of genetic instability and genomic features of HRD with BRCA1-associated and BRCA2-associated breast cancers with bi-allelic inactivation. Two of the 16 PALB2-associated breast cancers subjected to WES, however, lacked both bi-allelic inactivation of PALB2 and genomic features of HRD. Hence, we posit that molecular assays to identify bi-allelic inactivation of PALB2 and/or genomic features of HRD may aid in the selection of patients likely to benefit from HRD-directed therapies, including platinum-based chemotherapy and/or PARP inhibitors.
Methods
Cases
We included 24 invasive breast cancers from women with pathogenic PALB2 germline mutations. This study was approved by Memorial Sloan Kettering Cancer Center’s institutional review board (IRB) and by the local ethics committees/IRBs of the authors’ institutions. Written informed consents were obtained as required by the protocols approved by the IRBs/local ethics committees of the respective authors’ institutions. This study is in compliance with the Declaration of Helsinki. For 22 breast cancers, tissue samples were retrieved from the McGill University (Canada, n = 6), Cancer Research Malaysia/University Malaya (Malaysia, n = 5), the Kathleen Cuningham Foundation Consortium for research into Familial Breast Cancer (kConFab, Australia, n = 5), Memorial Sloan Kettering Cancer Center (MSKCC; New York, USA, n = 3), University of Eastern Finland (Finland, n = 2) and Charles University (Czech Republic, n = 1). Hematoxylin and eosin-stained tissue sections of the 22 breast cancers were reviewed by three pathologists (FCG, FP, and JSR-F). The genomics data of IDC53 were in part previously reported in Foo et al.9 In addition, the WES-derived mutational and clinico-pathologic data of two PALB2-associated breast cancers (IDC60 and IDC61; Table 1) were retrieved from the provisional TCGA breast cancer dataset at the Broad’s Institute firehose on 01/28/16.
Immunohistochemistry and fluorescence in situ hybridization (FISH)
ER and HER2 status were assessed by immunohistochemistry following American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guidelines.30 In addition, HER2 amplification was assessed in selected cases by fluorescence in situ hybridization (FISH) using PathVysion (Abbott) and/or HER2 IQFISH pharmDx (Dako), following the ASCO/CAP guidelines.31,32
DNA extraction
Eight-micrometer-thick sections from representative FFPE blocks were microdissected with a sterile needle under a stereomicroscope (Olympus) to ensure >80% of tumor cells. Genomic DNA was extracted from tumor and matched normal blood or saliva samples using the DNeasy Blood and Tissue Kit (Qiagen), and quantified using the Qubit Fluorometer (Life Technologies).
Massively parallel sequencing and bioinformatics analysis
DNA of tumor and matched normal samples was subjected to WES (n = 14) or MSK-IMPACT20 (n = 8), which targets all exons and selected introns of 410 cancer genes. Sequencing data analysis was performed as described previously.33,34 Read alignment and mutation calling was performed as described in the Supplementary Methods.35 CNAs and regions of LOH were defined using FACETS.36 In brief, homozygous deletions correspond to regions with an absolute copy number of zero, hemizygous losses are regions with absolute copy number lower than the average ploidy rounded to the nearest integer value. Low copy number gains are regions with absolute copy number greater than the average ploidy rounded to the nearest integer value, and high-level copy number amplifications are regions with absolute copy number in excess of four relative to the average ploidy rounded to the nearest integer value. Regions of LOH and homozygous deletions were manually reviewed using plots of Log2 ratios and B allele frequencies. The cancer cell fraction (CCF) of each mutation was inferred using ABSOLUTE (v1.0.6)37 and manually reviewed33,37,38 (Supplementary Methods).
Large-scale state transitions, indel length and mutational signatures
The presence of LSTs, representative of genomic scars indicative of HRD,39 was assessed in breast cancers subjected to WES. A LST score cut-off of 15 was adopted to classify breast cancers as LST-high (≥15) or LST-low (<15), as previously described.9,22,39 The length of indels was assessed in PALB2-associated breast cancers analyzed by WES, given that deletion sizes of ≥5 bp have been associated with defective HR-based repair.25 Mutational signatures40 were inferred from non-synonymous and silent somatic exonic SNVs (i) using deconstructSigs41 based on the set of 30 mutational signatures represented in COSMIC27 or (ii) on the 12 mutational signatures known to occur in breast cancers27 and (iii) using a non-negative matrix factorization algorithm (NMF)42 based on the 30 signatures from COSMIC,27 in samples with at least 30 somatic mutations, as previously described.15,22 The dominant mutational signature in each case was defined based on the consensus of at least two of the three methods.
Sanger sequencing validation
Selected somatic mutations with MAFs > 10%, including mutations affecting PIK3CA (n = 7), PALB2 (n = 4), TP53 (n = 4), and NOTCH3 (n = 2), were validated by Sanger sequencing (primer sequences in Supplementary Table 3). PCR amplification of genomic DNA and analyses were performed in duplicate.
Comparisons with breast cancers from TCGA
The mutation burden, mutation frequencies, CNAs and genomic features indicative of HRD of the PALB2-associated breast cancers were compared to those of non-BRCA1/2/PALB2-associated breast cancers with matched ER and HER2 status (n = 683), and to those of BRCA1 (n = 17) and BRCA2 (n = 16) breast cancers with bi-allelic inactivation from TCGA43 (Supplementary Methods).
Statistical analysis
Comparisons of the number of somatic mutations and LST scores, gene-level copy number states and mutational signatures between PALB2-associated breast cancers and non-BRCA1/2/PALB2-associated, BRCA1-associated and BRCA2-associated breast cancers were performed using the Mann–Whitney U test and Fisher’s exact test, respectively. To account for differences in sample sizes, a bootstrap resampling analysis was performed (Supplementary Methods).
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
WES sequencing data (supporting Fig. 1–5, Table 1, Supplementary Figs. 2–6 and supplementary tables 1–5) and MSK-IMPACT sequencing data (supporting Figs. 1, 2, 4 and 5, Table 1, supplementary Figs. 2, 4 and 5 and supplementary tables 1–5) generated during this study, can be accessed from cBioPortal (https://identifiers.org/cbioportal:brca_msk_li_2019). TCGA Breast Cancer sequencing data (supporting Figs. 4 and 5, supplementary Figs. 4–6 and supplementary table 5) used in this study, can be accessed from cBioPortal (https://identifiers.org/cbioportal:brca_tcga_pan_can_atlas_2018) or from the related publication https://doi.org/10.1016/j.cell.2018.02.060. Additional data supporting supplementary table 4 can be accessed from table 2 and supplementary table 1 of the related publication: https://doi.org/10.1002/path.5055. The data generated and analyzed during this study are described in the following data record: https://doi.org/10.6084/m9.figshare.8138912.
Change history
19 November 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Xia, B. et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 22, 719–729 (2006).
Reid, S. et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 39, 162–164 (2007).
Kanchi, K. L. et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat. Commun. 5, 3156 (2014).
Antoniou, A. C. et al. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 371, 497–506 (2014).
Takeuchi, S., Doi, M., Ikari, N., Yamamoto, M. & Furukawa, T. Mutations in BRCA1, BRCA2, and PALB2, and a panel of 50 cancer-associated genes in pancreatic ductal adenocarcinoma. Sci. Rep. 8, 8105 (2018).
Chen, S. & Parmigiani, G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 25, 1329–1333 (2007).
Cybulski, C. et al. Clinical outcomes in women with breast cancer and a PALB2 mutation: a prospective cohort analysis. Lancet Oncol. 16, 638–644 (2015).
Nikkila, J. et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat. Commun. 4, 2578 (2013).
Foo, T. K. et al. Compromised BRCA1-PALB2 interaction is associated with breast cancer risk. Oncogene 36, 4161–4170 (2017).
Isaac, D., Karapetyan, L., Tamkus, D. Association of germline PALB2 mutation and response to platinum-based chemotherapy in metastatic breast cancer: a case series. JCO Precision Oncol. 2, 1–5 (2018).
Knudson, A. G. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer 1, 157–162 (2001).
Lee, J. E. A. et al. Molecular analysis of PALB2-associated breast cancers. J. Pathol. 245, 53–60 (2018).
Potapova, A., Hoffman, A. M., Godwin, A. K., Al-Saleem, T. & Cairns, P. Promoter hypermethylation of the PALB2 susceptibility gene in inherited and sporadic breast and ovarian cancer. Cancer Res. 68, 998–1002 (2008).
Poumpouridou, N. et al. Development and validation of molecular methodologies to assess PALB2 expression in sporadic breast cancer. Clin. Biochem. 49, 253–259 (2016).
Riaz, N. et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat. Commun. 8, 857 (2017).
Foulkes, W. D. et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 9, R83 (2007).
Erkko, H. et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature 446, 316–319 (2007).
Ramus S. J. et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J. Natl Cancer Inst. 107, pii: djv214 (2015).
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Cheng, D. T. et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Honrado, E., Osorio, A., Palacios, J. & Benitez, J. Pathology and gene expression of hereditary breast tumors associated with BRCA1, BRCA2 and CHEK2 gene mutations. Oncogene 25, 5837–5845 (2006).
Weigelt, B. et al. The landscape of somatic genetic alterations in breast cancers from ATM germline mutation carriers. J. Natl. Cancer Inst. 110, 1030–1034 (2018).
Turner, N. C. & Reis-Filho, J. S. Tackling the diversity of triple-negative breast cancer. Clin. Cancer Res. 19, 6380–6388 (2013).
Polak, P. et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 49, 1476–1486 (2017).
Alexandrov, L. et al. The repertoire of mutational signatures in human cancer. bioRxiv. https://doi.org/10.1101/322859 (2018).
Telli, M. L. et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 22, 3764–3773 (2016).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Maxwell, K. N. et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat. Commun. 8, 319 (2017).
Mutter, R. W. et al. Bi-allelic alterations in DNA repair genes underpin homologous recombination DNA repair defects in breast cancer. J. Pathol. 242, 165–177 (2017).
Hammond, M. E. et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 28, 2784–2795 (2010).
Wolff, A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 31, 3997–4013 (2013).
Wolff, A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab Med. 138, 241–256 (2014).
Ng, C. K. Y. et al. The landscape of somatic genetic alterations in metaplastic breast carcinomas. Clin. Cancer Res. 23, 3859–3870 (2017).
Geyer, F. C. et al. Recurrent hotspot mutations in HRAS Q61 and PI3K-AKT pathway genes as drivers of breast adenomyoepitheliomas. Nat. Commun. 9, 1816 (2018).
Li, A. et al. Metadata supporting data files of the related manuscript: homologous recombination DNA repair defects in PALB2-associated breast cancers. figshare https://doi.org/10.6084/m9.figshare.8138912 (2019).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Landau, D. A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013).
Popova, T. et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 72, 5454–5462 (2012).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016).
Gaujoux, R. & Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinforma. 11, 367 (2010).
Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385 e318 (2018).
Acknowledgements
The authors thank the PALB2 interest group for their contributions to this study. The authors thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. Research reported in this paper was supported in part by the Breast Cancer Research Foundation and the Sarah Jenkins Fund, a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant No. P30CA008748; MSK), a grant of the Ministry of Health of the Czech Republic (NV15-29959A), Charles University projects PROGRES Q28/LF1 and SVV2019/260367, an HIR Grant UM.C/HlR/MOHE/06 from the Ministry of Higher Education, Malaysia, and the National Health and Medical Research Council, Australia (NHMRC, Project Grant APP1029974). kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania, and South Australia, and the Cancer Foundation of Western Australia. W.D.F. was funded in part by Susan G Komen. A.L. was supported by the China Scholarship Council. T.N.-D. is an Early Career Fellow of the National Breast Cancer Foundation and M.S. is a NHMRC Senior Research Fellow of the National Health and Medical Research Council. M.T. was funded by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, Addenbrooke’s Hospital and European Union Seventh Framework Program (2007–2013)/European Research Council (310018). S.P. was supported by the Swiss National Science Foundation (Ambizione grant number: PZ00P3_168165). J.S.R-F. is partly funded by the Breast Cancer Research Foundation and Britta Weigelt by Cycle for Survival.
Author information
Authors and Affiliations
Consortia
Contributions
B.W., S.H.T, M.T., W.D.F. and J.S.R.-F. conceived the study. kConFab Investigators, Z.R., T.N.-D., P.P., C.T., T.T., K.R., S.E.B., A.M., R.W., M.J., P.R., B.X., P.S.N, L.-M.L. and M.S. provided samples. F.C.G., R.B. and J.S.R.-F. reviewed the cases. P.B., J.Y.L., P.S., D.N.B., S.S.K.L. and R.K. performed the bioinformatics analysis. A.L. F.C.G., P.B., J.Y.L., P.S., D.N.B., F.P., S.S.K.L., R.K., B.R., R.B., S.P., H.Y.W., J.R.L., R.G.-M., L.C., L.N., M.E.R. analyzed and interpreted the data. A.L., F.C.G., F.P., B.W. and J.S.R.-F. wrote the first paper, which was reviewed by all coauthors.
Corresponding author
Ethics declarations
Competing interests
M.E.R. reports consultancy fees from McKesson and AstraZeneca, and uncompensated consulting/advisory activities with Merck and Pfizer. J.S.R.-F. reports personal/consultancy fees from VolitionRx, Page.AI, Goldman Sachs, Grail, Ventana Medical Systems, Invicro, and Genentech, outside the scope of the submitted work. All remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, A., Geyer, F.C., Blecua, P. et al. Homologous recombination DNA repair defects in PALB2-associated breast cancers. npj Breast Cancer 5, 23 (2019). https://doi.org/10.1038/s41523-019-0115-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-019-0115-9
This article is cited by
-
PALB2-mutated human mammary cells display a broad spectrum of morphological and functional abnormalities induced by increased TGFβ signaling
Cellular and Molecular Life Sciences (2024)
-
Discovery of a small-molecule inhibitor that traps Polθ on DNA and synergizes with PARP inhibitors
Nature Communications (2024)
-
DNA repair pathways in breast cancer: from mechanisms to clinical applications
Breast Cancer Research and Treatment (2023)
-
Validation of genomic and transcriptomic models of homologous recombination deficiency in a real-world pan-cancer cohort
BMC Cancer (2022)
-
Functional RECAP (REpair CAPacity) assay identifies homologous recombination deficiency undetected by DNA-based BRCAness tests
Oncogene (2022)