Abstract
The fitness consequences of inbreeding and the individual behaviors that prevent its detrimental effects can be challenging to document in wild populations. Here, we use field and molecular data from a 17-year study of banner-tailed kangaroo rats (Dipodomys spectabilis) to quantify the relationship between inbreeding, mate kinship, and lifetime reproductive success. Using a pedigree that was reconstructed using genetic and field data within a Bayesian framework (median probability of parental assignment = 0.92, mean pedigree depth = 6 generations), we estimated both inbreeding coefficients and kinship between individuals that produced offspring (mean inbreeding coefficient = 0.07, mean mate kinship = 0.08). We also used the pedigree, in combination with census data, to generate a series of fitness estimates, ranging from survival to reproductive maturity to lifetime reproductive success. We found that the population’s inbreeding load was low to moderate (0.98–4.66 haploid lethal equivalents) and increased with the time frame over which fitness was estimated (lowest for survival to maturity, highest for adult-to-adult reproductive success). Fitness decreased with increasing inbreeding coefficients. For example, lifetime reproductive success was reduced by 24% for individuals with inbreeding coefficients greater than twice the population mean. Within full sibling pairs, the sibling with less-related mates produced an average of 30% more offspring over its lifetime. These data further illustrate that inbreeding can have a negative effect on lifetime reproductive success.
Similar content being viewed by others
Introduction
Predicting the fitness consequences of inbreeding depression, and how those consequences are mitigated by behavior, is important for understanding how small or threatened populations can persist and whether they can respond to environmental change (Ralls et al. 1988; Caballero et al. 2017). Within a population, the magnitude of inbreeding varies among individuals (Forstmeier et al. 2012; Nietlisbach et al. 2017), meaning that inbreeding depression in the population can be altered by individual behaviors that influence mate choice (Brooks and Endler 2001). However, the complex interplay between the accumulation of inbreeding load and behaviors that reduce it is not yet fully understood, especially in wild populations. Here, we evaluate the fitness consequences of inbreeding on lifetime reproductive success using a near-complete 17-year pedigree in the solitary, desert-dwelling banner-tailed kangaroo rat (Dipodomys spectabilis).
Banner-tailed kangaroo rats have small population sizes, suggesting that opportunities for inbreeding with close relatives are common (Jones et al. 1988). Breeding with close relatives can have negative fitness consequences (inbreeding depression) because such mating exposes inbreeding load (i.e., deleterious recessive mutations concealed in heterozygotes, sensu Morton et al. 1956). Two broad categories of behaviors could limit the negative fitness consequences of inbreeding: dispersal and mate choice. For example, in species where dispersal occurs often and over large distances, dispersal can reduce the likelihood of adults encountering and subsequently mating with relatives (Frame et al. 1979; Smith et al. 2017). In the banner-tailed kangaroo rat, however, many individuals fail to disperse and, for those that do, dispersal distances are short (Waser et al. 2006). As a result, related individuals tend to remain in close proximity to each other even after dispersal (Winters and Waser 2003).
In the absence of dispersal, behaviors that reduce the frequency of matings between related individuals may be favored (Pusey and Wolf 1996; Keller and Waller 2002). In wild populations, extra-pair copulations can increase the probability of mating with unrelated individuals (Foerster et al. 2003), and kin discrimination can enable direct avoidance of close inbreeding (Lehmann and Perrin 2003). In banner-tailed kangaroo rats, females often mate with multiple males, thereby reducing the mean inbreeding coefficient for their offspring (Waser and DeWoody 2006). Furthermore, kangaroo rats in this population avoid mating with familiar kin; for example, they rarely mate with maternal siblings from the same litter (Waser et al. 2012).
In the long term, the negative effects of inbreeding on fitness may also be influenced by direct selection against deleterious alleles. For example, when inbreeding load is substantial, the greatly reduced survival and near zero lifetime reproductive success of inbred individuals can reduce inbreeding load by removing recessive, deleterious mutations from the population (i.e., purging, Crnokrak and Barrett 2012; Hedrick and Garcia-Dorado 2016). Additionally, behavioral and purging mechanisms can interact; behaviors that reduce the frequency of matings between kin can limit the exposure of deleterious alleles and, thus, the rate of purging.
Here, we use a combination of mark-recapture data and existing multi-locus genotypes to reconstruct a pedigree for a small (mean number of adults = 63, range 22–151) population of banner-tailed kangaroo rats. Data come from 1754 individuals followed over 17 years (Waser and Hadfield 2011; Waser et al. 2013). We address four questions: 1. What is the population’s inbreeding load? 2. How much does an increased inbreeding coefficient reduce fitness, as measured by survival to maturity, lifespan, total number of juvenile offspring produced, or lifetime reproductive success? 3. How much does increased mate kinship reduce fitness, by the same measures?, and 4. How much does mate kinship affect fitness after controlling for differences in inbreeding coefficients? We answer these questions using detailed census data and estimates of inbreeding and kinship generated from a deep pedigree.
Materials and methods
Banner-tailed kangaroo rats are solitary, larder-hording granivores that typically occur in arid grasslands (Vorhies and Taylor 1922). Individuals typically live two years (range 1–6), breed seasonally, and reach sexual maturity at age one (Waser and Jones 1991). Females generally produce 1–3 pups in a single litter each year, although up to three litters may be produced in years of high productivity (Jones 1984). Kangaroo rats construct large mounds (1–3 meters in diameter) that protect them from predators and harsh abiotic conditions; these mounds also serve as vessels for seed storage (Kay and Whitford 1978). Because construction of new mounds is time and energy intensive (Best 1972), most young of the year attempt to take over empty existing mounds, particularly those in close proximity to their birth mound (Jones 1984). From 1990 to 2007, Waser et al. (Waser and Jones 1991; Skvarla et al. 2004; Sanderlin et al. 2012) conducted twice-annual monitoring of a meta-population of banner-tailed kangaroo rats located in southeastern Arizona [31°37’N, 109°15’W] (Sanderlin et al. 2012; Fig. 1). Monitoring occurred by placing three traps around each occupied mound on three consecutive nights (median capture probability for adults 98% and juveniles 93%; Skvarla et al. 2004). All individuals were uniquely marked with ear tags, and pinna biopsies were used as a DNA source. Trapped individuals were genotyped at nine polymorphic microsatellite loci (average number of alleles = 7, mean expected heterozygosity = 0.63; see Waser et al. 2006; Busch et al. 2009 for additional details).
Location of study site in southeast Arizona. Points depict the locations of all occupied mounds across the 17-year-study period. Mounds occurred around the perimeter of a volcanic cinder cone. Top right inset show site location in North America. Bottom right inset illustrates the study species, banner-tailed kangaroo rat (Dipodomys spectabilis)
In order to generate estimates of individual inbreeding and fitness, we reconstructed population pedigrees using microsatellite genotypes, trapping records, and the location of each individual’s mound with MasterBayes (Hadfield et al. 2006). We ran MasterBayes for 600,000 iterations, with a thinning interval of 250 and a burn-in of 100,000 iterations. We used flat priors for all parameters, and the allelic dropout and stochastic error rates were set to 0.01 (Waser and Hadfield 2011; Waser et al. 2013). Within the model, parental assignment probabilities were influenced by trapping location as well as by parent and offspring genotypes. Adult females trapped at or near a young animal’s first capture site were assumed to be more likely to be the mother, and males trapped near the mother were preferred as sires (Waser and DeWoody 2006; Waser and Hadfield 2011). These model assumptions were based on trapping, radiotracking, and spool-and-line tracking that demonstrated short dispersal distances and small home ranges, even during the breeding season (Steinwald et al. 2013). Furthermore, earlier CERVUS-based parentage analysis found that sires rarely lived >100 m from their mates (Marshall et al. 1998; Winters and Waser 2003), and genotyping of copulatory plugs demonstrated that females primarily mated with nearby males (McCreight et al. 2011).
We used the most-likely pedigree generated by MasterBayes to estimate an inbreeding coefficient for each individual and to estimate relatedness between breeding pairs. We input our pedigree into the R package pedigree (Coster 2013) to calculate each individual’s inbreeding coefficient, defined as the probability that it possessed two alleles that were identical by descent at a particular locus (Fig. 2). We estimated kinship – the probability that two randomly sampled alleles are identical by descent between two individuals (Fig. 2) – using the R package kinship2 (Therneau and Sinnwell 2015), and calculated the mean kinship between each individual and its mates.
Hypothetical pedigree displaying pedigree calculations. In this example, individuals N (female) and P (male) have different histories of inbreeding in their family lines, leading to different inbreeding coefficients (F). The kinship (k) between the two individuals is smaller than either of their inbreeding coefficients because they have only one known ancestor in common (individual c). All letters represent distinct individuals
In addition to estimates of inbreeding and kinship, we also generated four estimates of individual fitness. First, we used trapping records to determine individual lifespan (in years). Second, we considered survival from first trapping, which occurred within a month or two of weaning, to maturity, as this parallels a common approach in other studies of inbreeding in wild vertebrate populations (Nietlisbach et al. 2018). For each individual, we also generated two additional measures of fitness using the pedigree. For each adult, we determined the total number of offspring that survived long enough to be captured, which we hereafter refer to as “number of juvenile offspring”. For each adult we additionally determined the total number of offspring produced that survived to reproductive maturity, which we hereafter refer to as “lifetime reproductive success”.
When individuals were trapped as young of the year, we used year of trapping as the birth year. Relatively few animals were first trapped as adults after the first year of sampling (n = 10); we assumed these were 1-year-olds since, in earlier fall trapping of these populations, nearly all trapped individuals that lacked a tag were large juveniles (Jones 1988). Because mark-recapture data demonstrated that we captured nearly all adults during each census and that emigration rates were very low (Skvarla et al. 2004; Sanderlin et al. 2012) we assumed year of death was the last year an individual was trapped.
We also estimated the inbreeding load of our kangaroo rat population, quantified as the number of haploid lethal equivalents (Morton et al. 1956). Following Morton et al. (1956), lethal equivalents can be quantified as the slope of a regression comparing the inbreeding coefficient (predictor variable) to the log of an estimate of fitness (response variable). However, this model can be problematic when fitness estimates equal zero (Nietlisbach et al. 2018). Therefore, we quantified the number of lethal equivalents using a modified version of the Morton et al. (1956) equation, where the relationship between fitness, inbreeding coefficient, and inbreeding load is determined by fitting an exponential model (Kalinowski and Hedrick 1998). We chose this model because it is unbiased and, therefore, provides estimates of inbreeding loads that are comparable between populations (Nietlisbach et al. 2018). In addition, we fit a modification of the Kalinowski and Hedrick (1998) model that was designed to quantify the effects of purging (García-Dorado 2012). We fit both equations using nonlinear regressions. Regression coefficients were estimated using a Bayesian computation algorithm that approximates maximum likelihood (Nietlisbach et al. 2018) via the program PURGd (Garcia-Dorado et al. 2016). For each model, we repeated the fitting process 100 times and subsequently report the mean number of haploid lethal equivalents as well as the standard deviation (SD) around the mean. Finally, we used a bootstrap analysis (100 regression replicates, implemented in the program PURGd) to understand the effects of including purging on our estimates of inbreeding load compared to the model that did not include purging (Garcia-Dorado et al. 2016). We ran PURGd using default parameters (i.e., we allowed the program to compute the initial average fitness and inbreeding load and set the maximum initial fitness value to 1; Garcia-Dorado et al. 2016), our kangaroo rat pedigree, and our four measures of fitness: lifespan, survival from first trapping to maturity, number of juvenile offspring, and lifetime reproductive success.
We illustrated the relationship between individual fitness and inbreeding using generalized linear models in R 3.4.3 (R Core Team 2017). We included only individuals born prior to 2005 because some members of our last two cohorts were still alive at the end of our study. Because estimates of inbreeding can be error prone at small pedigree depths (Kardos et al. 2015), we also filtered by the number of generations of family history reconstructed for each individual; we limited our inferences to parents (i.e., individuals with at least one identified offspring at the time of first trapping) that had at least three generations of ancestors captured by the pedigree (i.e., the minimum pedigree depth was four generations, where all 16 great-grandparents were known).
We assessed the effects of inbreeding on fitness using a series of regressions. First, we used a generalized mixed-effects model to determine the relationship between the pedigree-estimated inbreeding coefficient (explanatory variable) and the number of years an individual survived (lifespan; response variable). In this and subsequent regressions, we used Poisson models with a log-link function, fit with a Laplace approximation, and treated the individual’s birth year as a random variable to account for the variance among years in resource availability. We used the R package lme4 to fit the model (Bates et al. 2014). We used similar mixed-effects models to estimate the effects of inbreeding coefficient on survival to maturity, number of juvenile offspring and lifetime reproductive success, and (as described below) to estimate effects of kinship on fitness. In all cases, we subsequently estimated the variation explained by all the models (marginal variance explained by fixed effects only R2M; conditional variance explained by fixed and random effects R2C) using the R package piecewiseSEM (Nakagawa and Schielzeth 2013; Lefcheck 2016).
To measure the effects of mate kinship on fitness, we compared the mean kinship between an individual and all of its mates to its lifetime reproductive success. We could not perform this analysis on a mating by mating basis, which may decrease some of the noise associated with averaging over multiple mating events, because we could not assign-specific juveniles to specific litters as the same two parents can have multiple litters per year (Winters and Waser 2003). An individual with a smaller mean kinship to its mates is one that mated with less-related individuals, on average, than an individual with a larger mean kinship to its mates. We ran our models using all individuals (regardless of sex) and after sub-setting our data by sex.
Finally, we used full-sibling pairs to understand the potential for kin discrimination and mate choice to influence fitness. Full sibling pairs have the same history of inbreeding in their family lines but may differ in relatedness to their mates. It is important to note that full siblings have identical pedigree-estimated inbreeding coefficients and have all of the same ancestors, but can differ in genome-wide estimates of inbreeding due to variation in Mendelian inheritance patterns (Knief et al. 2017). To the extent that increased mate kinship decreases offspring fitness, full siblings that differ more in mean mate kinship would be expected also to differ more in lifetime reproductive success. Thus, using full-sibling pairs can control, at least partially, for inbreeding history. We identified groups of full siblings, then computed two values: 1. difference in mean mate kinship (mean kinship between one sibling and all of its mates over a lifetime minus the mean kinship for the other sibling and all of its mates over a lifetime) and 2. relative fitness, estimated as both the number of juvenile offspring and lifetime reproductive success. Both values were computed with respect to the sibling with the larger mean mate kinship in the pair, so that if inbreeding depression exists, the expected difference in reproductive success would be negative. Mathematically, the difference in mean mate kinship equals
where s1 is the sibling with higher mean mate kinship, ks1 equals the kinship between s1 and mate m1 (summed from 1 to m1, all of s1’s mates), and ks2 equals the kinship between s2 and mate m2 (summed from 1 to m2, all of s2’s mates). Relative reproductive success equals
where Noffs2 equals the number of offspring produced by sibling 2 (the sibling with lower mean mate kinship) summed across all of its m2 mates and Noffs1 equals the number of offspring produced by sibling 1 (the sibling with higher mean mate kinship) summed across all of its m1 mates. Using these values, we then estimated the effect of differences in mate kinship on relative fitness using a linear regression and again treated birth year as a random variable. Greater differences in mate kinship should result in larger changes in fitness, and we expected a negative relationship in the presence of inbreeding depression. We interpreted the strength of this relationship as indicating the extent to which mate kinship could potentially influence fitness. In addition, we ran a second set of models that included the siblings’ pedigree-estimated inbreeding coefficient as an additional predictor. All of these linear-errors models were fitted by restricted maximum likelihood (Pinheiro et al. 2018).
Results
We identified both parents for 1441 individuals born between 1990 and 2007 (Supplementary Table 1) out of 1754 individuals born during this time frame (i.e., parents were identified for 82.1% of individuals) with a median, posterior probability of parental assignments of 0.92 (Figure S1). The resulting pedigree had a mean depth of 6 generations (Figure S2). Based on the reconstructed pedigree, we found that adults (both males and females) who mated successfully did so most often with 1–2 mates over their lifetime (Figure S3), and that lifetime reproductive success ranged from 0 to 12, although very few adults produced more than 1–2 offspring (Figure S4). For individuals that reproduced and met our pedigree depth cutoff criterion, the mean inbreeding coefficient was 0.07 and the mean mate kinship was 0.08 (Figure S5).
We quantified inbreeding load as the number of haploid lethal equivalents using our pedigree and our four measures of fitness. We found that inbreeding load ranged from 0.98 (survival from first trapping to maturity) to 4.66 (lifetime reproductive success) lethal equivalents (Table 1). Across all fitness measures, the number of lethal equivalents estimated when considering the effects of purging (i.e. the inbreeding-purging model, García-Dorado 2012) was not significantly different than the estimated number of lethal equivalents when purging was not included in the model (i.e. Kalinowski and Hedrick 1998). Furthermore, none of our estimates of purging rate differed significantly from zero (bootstrapped p-values all >0.05; Table 1).
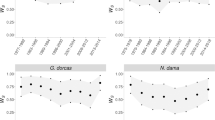
Fitness, whether approximated by lifespan, number of juvenile offspring or lifetime reproductive success, was negatively related to inbreeding. We found that an individual’s inbreeding coefficient (estimated from the pedigree) was negatively related to its lifespan (Table 2; Fig. 3). We also found a significant-negative relationship between an individual’s inbreeding coefficient and its lifetime number of juvenile offspring (Table 2; Fig. 4a). Similarly, an individual’s lifetime reproductive success was negatively related to its inbreeding coefficient (Table 2; Fig. 4c). Lifetime reproductive success for breeding individuals was reduced by 24% for individuals with an inbreeding coefficient greater than twice the population mean. Analyzing the two sexes separately, we found similar relationships between inbreeding and fitness in males and females (Figure S6-S8).
Relationship between pedigree-estimated inbreeding coefficient and lifespan. The line depicts the generalized linear regression line (birth year treated as random variable; Poisson errors; variance explained by fixed effects only). On average, individuals that were less inbred survived longer than individuals with higher inbreeding coefficients. (See Table 2 for coefficient estimates and other model results)
Pedigree estimates of fitness as a function of individual inbreeding coefficient a, c or mean kinship between an individual and all of its mates b, d. In a, b the y-axis depicts the number of juvenile offspring, whereas in c, d it represents lifetime reproductive success. In all plots, inbreeding and fitness estimates were compared using a generalized linear model with Poisson errors, treating parental birth year as a random variable. In each panel, the resulting regression line is plotted over the individual data points. (See Table 2 for coefficient estimates and other model results)
Across the population as a whole, we found that mean kinship between mates was not related to number of juvenile offspring (Table 2; Fig. 4b). Similarly, we found no significant relationship between mean mate kinship and lifetime reproductive success (Table 2; Fig. 4d). There was also no relationship, across the entire population, between mean mate kinship and number of offspring produced when using single-sex models (Figures S7 and S8).
In contrast, when we confined our analysis to full-sibling pairs, we found that higher kinship among mates resulted in decreased reproductive output. Siblings with mates of similar mean kinship produced similar numbers of offspring but, when siblings’ mean kinship to their mates differed, the sibling with the higher mean mate kinship produced significantly fewer juvenile offspring (Table 2; Fig. 5). An increase in mean mate kinship of 0.15 resulted in the production of 59% fewer juvenile offspring (Fig. 5a). Similarly, siblings with higher mean mate kinship had significantly lower lifetime reproductive success (Table 2). A 0.15 difference between siblings in mean mate kinship decreased lifetime reproductive success by 46% (Fig. 5b). Finally, models that included sibling inbreeding coefficient as an additional explanatory variable produced similar results, and inbreeding coefficient was not a significant predictor of relative fitness in these models for either the number of juvenile offspring or lifetime reproductive success (Tables S2-S3).
Difference in relative reproductive success between siblings within a full-sibling pair as a function of difference in mean mate kinship between the same siblings. In a, the y-axis shows the difference between siblings in number of juvenile offspring. In b, it denotes the difference in lifetime reproductive success. In both panels, reproductive success values are relative to the sibling with the larger mean mate kinship such that values <1 indicate that the sibling with the more closely related mates had fewer offspring. The dashed horizontal line indicates the expectation if kinship has no effect on reproductive success. The blue line illustrates the results of a generalized mixed-effect linear model, where birth year was modeled as a random variable. (See Table 2 for coefficient estimates and other model results)
Discussion
We used a 17-year data set from wild banner-tailed kangaroo rats and identified inbreeding depression as the negative relationship between an individual’s pedigree-estimated inbreeding coefficient and fitness. Results of previous studies of mammalian populations where the effects of inbreeding were derived from reconstructed pedigrees have been mixed. Some studies have reported non-significant or weak negative relationships between fitness and pedigree-estimated inbreeding coefficients (Keane et al. 1996; Huisman et al. 2016; Wells et al. 2018), whereas others identified significant and stronger negative relationships (Liberg et al. 2005; Rioux-Paquette et al. 2011; Walling et al. 2011; Nielsen et al. 2012). The differences between these studies may reflect biological differences between species, or may be due to the relatively lower power of pedigrees to detect inbreeding depression compared to genomic data (Kardos et al. 2015). Here, we found a significant-negative relationship between pedigree-estimated inbreeding coefficient and fitness, where fitness components were measured either in terms of survival or lifetime reproductive success (Figs. 3 and 4).
Across all of our fitness measures, our kangaroo rat population was characterized by a low to moderate inbreeding load (Table 2). Recently, Nietlisbach et al. (2018) provided updated estimates of haploid lethal equivalents from previously analyzed populations that are unbiased and, therefore, suitable for comparison across populations. Although available data are scarce, our estimates were typically near the lower end of the range reported for other wild populations using similar fitness measures. For example, our estimate of kangaroo rat inbreeding load based on survival from first trapping to maturity (0.98 lethal equivalents) was most directly comparable to estimates from other species based on individuals followed from fledging to recruitment. These range from 0.0 lethal equivalents (medium ground finch) to 7.5 lethal equivalents (collared flycatcher) (Nietlisbach et al. 2018). Our estimate of inbreeding load based on lifetime reproductive success (4.6 lethal equivalents) is far lower than the only other such estimate reported by Nietlisbach et al. (2018), 24.6 lethal equivalents for song sparrows. To our knowledge, no values based on lifetime reproductive success are available from other wild mammals, but comparable estimates based on less than the complete life cycle are available from golden lion tamarin (2.8 lethal equivalents), gray wolf (3.0 lethal equivalents), and red deer (4.4 lethal equivalents) (Nietlisbach et al. 2018).
Behavioral mechanisms, such as those that reduce the relatedness between mates, can reduce the negative fitness consequences associated with inbreeding by reducing the inbreeding coefficient of any resulting offspring (Pusey and Wolf 1996). In this population, we found that when we compared the number of offspring produced by full siblings, the sibling with mates more closely related to itself produced fewer offspring (Fig. 5); over its lifetime, the sibling with less-related mates produced an average of 30% more offspring that survived to maturity. It is important to note that the relationship between mean mate kinship and reduced fitness was found only when we compared full siblings and not all individuals (cf. Fig. 4b, d and Fig. 5). In the population at large, the impact of mating with close relatives on reproductive success may have been masked by other temporally and spatially mediated factors that influence fitness (e.g., weather, predator pressure, resource availability, maternal care). Restricting the analysis to full siblings indicates the magnitude of the fitness advantage that individual kangaroo rats might gain through behaviors that reduce the probability of mating with close relatives, thereby avoiding the effects of deleterious alleles that are more likely to be expressed in homozygous offspring.
Despite our population’s relatively low inbreeding load, our estimates of the current rates of purging do not differ significantly from zero (Table 1). This result may suggest that our data set has low power to detect purging or that the purging occurred before the beginning of this study. It also highlights the need for a better understanding of behaviors, like mate choice based on kin discrimination, which can influence the exposure of deleterious recessives to selection. Banner-tailed and other species of kangaroo rats can distinguish neighbors, related or not, from unfamiliar individuals using olfactory or acoustic signals (Randall 1987; Murdock and Randall 2001). In addition, previous parentage analyses demonstrated that in our population, mother–son pairings virtually never occur, whereas father–daughter pairings are almost as common as would be expected by chance given the spatial distribution of parents and their adult offspring (Waser et al. 2012). Similarly, offspring of paternal half-siblings occur as often as would be expected if females mated randomly with regard to this relationship, whereas offspring of maternal half-siblings are rare. Because young kangaroo rats grow up in a mound with their mother and maternal siblings, but not their father or paternal half-siblings, the observed deficit of pairings with close maternal relatives is consistent with the idea that kangaroo rats discriminate against mating with recognizable kin (Waser et al. 2012).
Many vertebrates exhibit similar forms of kin discrimination based on association and familiarity (Tang-Martinez 2001), and in some populations, mate choice based on kin discrimination likely has a heritable basis (Jennions and Petrie 1995; Tregenza and Wedell 2000; Tenesa et al. 2015; Svensson et al. 2017). Familiarity-based kin discrimination should prevent matings with close maternal kin, thereby reducing the effectiveness of purging. But the impact of this form of mate choice on inbreeding load may vary among species. In particular, in this and many other species, paternal kin and more distant relatives are not “familiar” to an individual, meaning that not all inbred matings are prevented. To our knowledge, the potential effects of familiarity-based kin discrimination on estimates of inbreeding load have not been explored.
Small populations, especially those whose members tend to disperse short distances, are often assumed to be highly vulnerable to the detrimental effects of inbreeding. Long-term data from wild populations are, however, rare. Here, we have used a 17-year pedigree from one such population. Although pedigree-based estimates of inbreeding coefficients can have lower resolution than those based on genomic techniques (Nietlisbach et al. 2018), the ability of MasterBayes to reconstruct the pedigree and our ability to sample essentially all individuals over multiple generations helped to alleviate this constraint in our study. Using the pedigree, we were able to show that the fitness costs of inbreeding, measured in terms of survival or reproductive success, were substantial, reflecting the low to moderate inbreeding load documented in this small, dispersal-limited population.
Data archiving
The complete pedigree, which includes all parental assignments, is available in Supplementary Table 1. All analysis code is available via GitHub at https://github.com/jwillou/krat_inbreeding and https://github.com/ChristieLab/KangarooRat_inbreeding.
References
Bates D, Mächler M, Bolker B, Walker S (2014) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48
Best TL (1972) Mound development by a pioneer population of the banner-tailed kangaroo rat, Dipodomys spectabilis baileyi Goldman, in Eastern New Mexico. Am Midl Nat 87:201–206
Brooks R, Endler JA (2001) Female guppies agree to differ: phenotypic and genetic variation in mate-choice behavior and the consequences for sexual selection. Evolution 55:1644–1655
Busch JD, Waser PM, Dewoody JA (2009) The influence of density and sex on patterns of fine-scale genetic structure. Evolution 63:2302–2314
Caballero A, Bravo I, Wang J (2017) Inbreeding load and purging: implications for the short-term survival and the conservation management of small populations. Heredity 118:177–185
Coster A (2013). pedigree: Pedigree functions. R package version 1.6.4. https://CRAN.R-project.org/package=kinship2
Crnokrak P, Barrett SCH (2012) Purging the genetic load: a review of the experimental evidence. Evolution 56:2347–2358
Foerster K, Delhey K, Johnsen A, Lifjeld JT, Kempenaers B (2003) Females increase offspring heterozygosity and fitness through extra-pair matings. Nature 425:714–717
Forstmeier W, Schielzeth H, Mueller JC, Ellegren H, Kempenaers B (2012) Heterozygosity-fitness correlations in zebra finches: microsatellite markers can be better than their reputation. Mol Ecol 21:3237–3249
Frame LH, Malcolm JR, Frame GW, Van Lawick H (1979) Social organization of African wild dogs (Lycaon pictus) on the Serengeti Plains, Tanzania 1967–1978. Z Tierpsychol 50:225–249
García-Dorado A (2012) Understanding and predicting the fitness decline of shrunk populations: inbreeding, purging, mutation, and standard selection. Genetics 190:1461–1476
Garcia-Dorado A, Wang J, Lopez-Cortegano E (2016) Predictive model and software for inbreeding-purging analysis of pedigreed populations. G3 6:3593–3601
Hadfield JD, Richardson DS, Burke T (2006) Towards unbiased parentage assignment: combining genetic, behavioural and spatial data in a Bayesian framework. Mol Ecol 15:3715–3730
Hedrick PW, Garcia-Dorado A (2016) Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol Evol 31:940–952
Huisman J, Kruuk LEB, Ellis PA, Clutton-Brock T, Pemberton JM (2016) Inbreeding depression across the lifespan in a wild mammal population. Proc Natl Acad Sci USA 113:3585–90
Jennions MD, Petrie M (1995) Variation in mate choice and mating preferences: a review of causes and consequences. Biol Rev Camb Philos Soc 70:359–360
Jones WT (1984) Natal philopatry in bannertailed kangaroo rats. Behav Ecol Sociobiol 15:151–155
Jones WT (1988) Density-related changes in survival of philopatric and dispersing kangaroo rats. Ecology 69:1474–1478
Jones WT, Waser PM, Elliott LF, Link NE, Bush BB (1988) Philopatry, dispersal, and habitat saturation in the banner-tailed kangaroo rat. Dipodomys Spectabilis Ecol 69:1466–1473
Kalinowski ST, Hedrick PW (1998) An improved method for estimating inbreeding depression in pedigrees. Zoo Biol 17:481–497
Kardos M, Luikart G, Allendorf FW (2015) Measuring individual inbreeding in the age of genomics: marker-based measures are better than pedigrees. Heredity 115:63–72
Kay FR, Whitford WG (1978) The burrow environment of the banner-tailed kangaroo rat, Dipodomys spectabilis, in Southcentral New Mexico. Am Midl Nat 99:270–279
Keane B, Creel SR, Waser PM (1996) No evidence of inbreeding avoidance or inbreeding depression in a social carnivore. Behav Ecol 7:480–489
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Knief U, Kempenaers B, Forstmeier W (2017) Meiotic recombination shapes precision of pedigree- and marker-based estimates of inbreeding. Heredity 118:239–248
Lefcheck JS (2016) piecewiseSEM: Piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods Ecol Evol 7:573–579
Lehmann L, Perrin N (2003) Inbreeding avoidance through kin recognition: choosy females boost male dispersal. Am Nat 162:638–652
Liberg O, Andrén H, Pedersen H-C, Sand H, Sejberg D, Wabakken P et al. (2005) Severe inbreeding depression in a wild wolf (Canis lupus) population. Biol Lett 1:17–20
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
McCreight JC, Dewoody JA, Waser PM (2011) DNA from copulatory plugs can give insights into sexual selection. J Zool 284:300–304
Morton NE, Crow JF, Muller HJ (1956) An estimate of the mutational damage in man from data on consanguineous marriages. Proc Natl Acad Sci USA 42:855–863
Murdock HG, Randall JA (2001) Olfactory communication and neighbor recognition in giant kangaroo rats. Ethology 107:149–160
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142
Nielsen JF, English S, Goodall-Copestake WP, Wang J, Walling CA, Bateman AW et al. (2012) Inbreeding and inbreeding depression of early life traits in a cooperative mammal. Mol Ecol 21:2788–2804
Nietlisbach P, Keller LF, Camenisch G, Guillaume F, Arcese P, Reid JM et al. (2017) Pedigree-based inbreeding coefficient explains more variation in fitness than heterozygosity at 160 microsatellites in a wild bird population. Proc R Soc B Biol Sci 284:20162763
Nietlisbach P, Muff S, Reid JM, Whitlock MC, Keller LF (2018) Nonequivalent lethal equivalents: odels and inbreeding metrics for unbiased estimation of inbreeding load. Evol Appl 12:266–279
Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team (2018) nlme: linear and nonlinear mixed effects models. R package version 3.1-131.1, https://CRAN.R-project.org/package=nlme
Pusey A, Wolf M (1996) Inbreeding avoidance in animals. Trends Ecol Evol 11:201–206
R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/
Ralls K, Ballou JD, Templeton A (1988) Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv Biol 2:185–193
Randall JA (1987) Sandbathing as a territorial scent-mark in the bannertail kangaroo rat. Dipodomys spectabilis Anim Behav 35:426–434
Rioux-Paquette E, Festa-Bianchet M, Coltman DW (2011) Sex-differential effects of inbreeding on overwinter survival, birth date and mass of bighorn lambs. J Evol Biol 24:121–131
Sanderlin JS, Waser PM, Hines JE, Nichols JD (2012) On valuing patches: estimating contributions to metapopulation growth with reverse-time capture-recapture modelling. Proc R Soc B Biol Sci 279:480–488
Skvarla JL, Nichols JD, Hines JE, Waser PM (2004) Modeling interpopulation dispersal by banner-tailed kangaroo rats. Ecology 85:2737–2746
Smith JE, Lehmann KDS, Montgomery TM, Strauss ED, Holekamp KE (2017) Insights from long-term field studies of mammalian carnivores. J Mammal 98:631–641
Steinwald MC, Swanson BJ, Doyle JM, Waser PM (2013) Female mobility and the mating system of the banner-tailed kangaroo rat (Dipodomys spectabilis). J Mammal 94:1258–1265
Svensson O, Woodhouse K, Van Oosterhout C, Smith A, Turner GF, Seehausen O (2017) The genetics of mate preferences in hybrids between two young and sympatric Lake Victoria cichlid species. Proc R Soc B 284:20162332
Tang-Martinez Z (2001) The mechanisms of kin discrimination and the evolution of kin recognition in vertebrates: a critical re-evaluation. Behav Process 53:21–40
Tenesa A, Rawlik K, Navarro P, Canela-Xandri O (2015) Genetic determination of height-mediated mate choice. Genome Biol 16:269
Therneau TM, Sinnwell J (2015) kinship2: Pedigree Functions. R package version 1.4. https://CRAN.R-project.org/package=pedigree
Tregenza T, Wedell N (2000) Genetic compatibility, mate choice and patterns of parentage: invited review. Mol Ecol 9:1013–1027
Vorhies CT, Taylor WP (1922) Life history of the kangaroo rat, Dipodomys spectabilis spectabilis (Merriam). United States Dep Agric 1091:1–40
Walling CA, Nussey DH, Morris A, Clutton-Brock TH, Kruuk LE, Pemberton JM (2011) Inbreeding depression in red deer calves. BMC Evol Biol 11:318
Waser PM, Berning ML, Pfeifer A (2012) Mechanisms of kin discrimination inferred from pedigrees and the spatial distribution of mates. Mol Ecol 21:554–561
Waser PM, Busch JD, McCormick CR, DeWoody JA (2006) Parentage analysis detects cryptic precapture dispersal in a philopatric rodent. Mol Ecol 15:1929–1937
Waser PM, DeWoody JA (2006) Multiple paternity in a philopatric rodent: the interaction of competition and choice. Behav Ecol 17:971–978
Waser PM, Hadfield JD (2011) How much can parentage analyses tell us about precapture dispersal? Mol Ecol 20:1277–1288
Waser PM, Jones WT (1991) Survival and reproductive effort in banner-tailed kangaroo rats. Ecology 72:771–777
Waser PM, Nichols KM, Hadfield JD (2013) Fitness consequences of dispersal: is leaving home the best of a bad lot? Ecology 94:1287–1295
Wells DA, Cant MA, Nichols HJ, Hoffman JI (2018) A high-quality pedigree and genetic markers both reveal inbreeding depression for quality but not survival in a cooperative mammal. Mol Ecol 27:2271–2288
Winters JB, Waser PM (2003) Gene dispersal and outbreeding in a philopatric mammal. Mol Ecol 12:2251–2259
Acknowledgements
We thank our editor and three anonymous reviewers for providing comments and suggestions that substantially improved our work. We also thank A. Harder, M. Martinez, C. Schraidt, M. Sparks, and X. Yin for helpful comments on previous version of this manuscript, J. Hadfield for assistance with MasterBayes, and P Nietlisbach for insightful correspondence and suggestions regarding lethal equivalents. J.R.W. was partially supported by a fellowship through the Purdue Forestry and Natural Resources postdoctoral program. This research was funded by support to M.R.C. from the Purdue Department of Forestry and Natural Resources and the Department of Biological Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Willoughby, J.R., Waser, P.M., Brüniche-Olsen, A. et al. Inbreeding load and inbreeding depression estimated from lifetime reproductive success in a small, dispersal-limited population. Heredity 123, 192–201 (2019). https://doi.org/10.1038/s41437-019-0197-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-019-0197-z