Abstract
Marine-derived bacteria are a prolific source of a wide range of structurally diverse natural products. This review, dedicated to Professor William Fenical, begins by showcasing many seminal discoveries made at the University of California San Diego from marine-derived actinomycetes. Discussed early on is the 20-year journey of discovery and advancement of the seminal actinomycetes natural product salinosporamide A into Phase III anticancer clinical trials. There are many fascinating parallels discussed that were gleaned from the comparative literature of marine sponge, tunicate, and bacteria-derived natural products. Identifying bacterial biosynthetic machinery housed in sponge and tunicate holobionts through both culture-independent and culture-dependent approaches is another important and expanding subject that is analyzed. Work reviewed herein also evaluates the hypotheses that many marine invertebrate-derived natural products are biosynthesised by associated or symbiotic bacteria. The insights and outcomes from metagenomic sequencing and synthetic biology to expand molecule discovery continue to provide exciting outcomes and they are predicted to be the source of the next generation of novel marine natural product chemical scaffolds.
Similar content being viewed by others
Introduction
A brief sketch of seminal discoveries by Professor Fenical that set the stage for exploring bioactive substances from marine-derived bacteria
It has become increasingly evident that there is significant overlap between the biosynthetic machinery of marine-derived bacteria vs. that of chemically prolific invertebrates, especially sponges and tunicates. A continuing stream of statements in both primary research papers and reviews outline the hypothesis that many invertebrate-derived compounds are seemingly produced by the action of an invertebrate microbiome. Thus, microbial symbionts may be critical to the production of many marine invertebrate natural products [1]. Relevant to this possibility are two significant findings. In 2015, a family of complex alkaloids, containing three tetrahydroisoquinoline moieties, originally isolated from the tunicate Ecteinascidia turbinate, was eventually found to be produced by the unculturable bacterial endosymbiont Candidatus Endoecteinascidia frumentensis, obtained directly from metagenomic DNA [2]. Similarly, in 2017 it was noted that the unculturable and ubiquitous endosymbiont Candidatus Entotheonella detected in sponges including Theonella swinhoei (themselves a source of a diverse set of molecular structures) possessed biosynthetic richness akin to that of soil actinomycetes [3]. Thus, building a broad foundation on the biosynthetic capabilities of unique libraries of marine-derived actinomycetes is a requisite for catalyzing future research to gain a firm understanding about overlapping invertebrate/bacterial biosynthetic machinery.
The overall goal in this review is to trace selected examples of intersections in the scaffolds (i.e., biosynthetic pathways) of complex small molecules from marine sponges and tunicates vs. marine-derived bacteria. In this section, our focus is on fundamentals of marine-derived actinomycetes and discussions about invertebrates will come later. An obscure study published in 1959 illustrated that dozens of marine-derived actinomycetes, as obligate halophytes, could be isolated, maintained in stable culture, and further studied [4]. However, for years there was virtually no follow-up work by others especially in terms of marine natural product compound discovery. As will be discussed next, many years passed until this situation changed. The path forward involved challenging risk-taking research at the University of California San Diego (UCSD) beginning in the early 1990s. Initially it took almost a decade of trial-and-error investigations to achieve the first notable outcomes [5]. A selection of some of the most striking results is highlighted by the cluster of eight structures collected in Fig. 1.
It is interesting to note that at UCSD Professor Fenical often asked the question, “where are the new horizons in marine bioorganic chemistry?” [6]. We deem that the selected structures and accompanying annotations shown in Fig. 1 provide some answers to this crisp question and are relevant to the discussions contained in the latter sections of this review. The contents of Fig. 1 span the time window 1999–2020. We invite all readers to examine each structural drawing, then ponder their significance (shown in each panel), and gain insights on how this assemblage has catalyzed the defining and subsequently expanding fundamental understanding of the chemistry and biology of marine actinomycete bacteria.
Here is a brief synopsis of selected inspirational molecules discovered at UCSD from actinomycetes. Cyclomarin (1), the first entry in Fig. 1, embodies a rather complex scaffold in terms of atom count (C56H82N8O11), chiral centers (12), and molecular weight (MW = 1043) [7]. It has bioactivity against organisms that cause malaria and tuberculosis is also significant. The Phase III anticancer candidate salinosporamide A (2) (Sal A, aka NPI-0052, marizomib) continuously isolated from Salinispora tropica in good yields and recently isolated in very low yields from S. arenicola is a compound that remains of high value more than two decades after its discovery (see Fig. 2 for a timeline) [8]. The sporolides A (3) and B, also from S. tropica, have a fascinating biogenesis including the involvement of a para-benzyne intermediate [9]. The six MRSA active marinopyrroles headed by the axial chiral atropisomer (−)-marinopyrrole A (4) are under intense study by many labs and the (±) form called maritoclax is commercially available [10]. At first glance ammosamide B (5), a relatively small achiral pigment (MW = 291) discovered in 2009 might seem unimportant, however, ammosamide A and B (5) possess potent activity against HCT-116 cancer cells (IC50 = 320 nM) [11]. Also, unlocking the mysteries about their biosynthesis is stretching molecular genetics tools to yield new hypotheses for the biosynthetic pathways of these and other amino acid containing natural products [12]. (+)-Merochlorin A (6) is a novel MRSA active tetracyclic chlorinated merosesquiterpene [13]. Even though X-ray analysis provided relative configuration assignments, it took an additional 7 years to finalize the absolute configurations of the members of this family through total synthesis [14]. (−)-Anthracimycin (7) is an antibiotic whose name celebrates its activity against Bacillus anthracis (MIC = 0.03 µg/mL) the anthrax bioterrorism weapon [15]. The complete structure of neaumycin B (8), a spectacularly potent cytotoxin (IC50 = 0.07 pM) against U87 human glioblastoma, was recently deduced by the UCSD team and updated two incomplete previous reports by others [16]. Interestingly, several actinomycetes strains are a source of this unique spiroketal containing polyketide.
The salinosporamide story—from a marine actinomycete-derived natural product to clinical trials
There are only a few marine natural products that have progressed through the advanced stages of clinical trials or gained FDA approval for therapeutic use. Sal A (2) (aka NPI-0052 or marizomib) [8] is an important member of this select group (see Fig. 3) because of its recent (2020) entry into Phase III clinical trials [17]. “Sal A” (structure shown in Fig. 1 with selected background information) is produced from a surprisingly complex biosynthetic pathway. Shown in Fig. 2 is the 20-year timeline from the isolation and characterization of 2 from a salt-water obligate marine actinomycete, Salinispora (strain CNB-392) to its entry into Phase III anticancer clinical trials.
We will not reiterate the many details shown in Fig. 2 but will instead focus on some additional factors. (1) “Sal A” (2) was initially targeted for additional SAR, biosynthetic, and experimental therapeutic investigations. This was motivated by the potent, selective results in the NCI’s 60-cell-line panel with a mean GI50 < 10 nM and greater than a 4 log LC50 differential between resistant and susceptible cell lines. The greatest potency of 2 was observed against NCI-H226 non-small cell lung cancer, SF-539 brain tumor, SK-MEL-28 melanoma, and MDA-MB-435 melanoma all with LC50 values <10 nM [18]. (2) In 2005, a public (UCSD)–private partnership (Nereus) successfully launched preclinical evaluation culminating an Investigational New Drug application to initiate clinical testing on 2 [19, 20]. (3) Early on, the anticancer molecular target was identified when 2 was tested against purified 20S proteasome, it inhibited proteasomal chymotrypsin-like proteolytic activity with an IC50 value of 1.3 nM. This action is similar to that of omuralide [21], a known inhibitor of proteasome which has structural similarities but important structural deficiencies for bioactivity vs. 2. (4) Even though many, somewhat lengthy, total syntheses have been published [22], a scale-up saline fermentation is being used to provide natural 2 from strain NPS21184 via a current good manufacturing practice scheme [18]. (5) An important encouraging finding is that 2 can cross the blood-brain barrier, motivating the Phase III clinical trials in patients with glioblastoma [17]. In summary, an unusual marine-derived shallow and deep-water salt obligate actinomycete has provided 2 as a promising new treatment for glioblastoma, the cancer that killed Sen. John McCain.
Comments on the recent drug development progress for bioactive marine natural products derived from sponges, tunicates, or bacteria
In section “The salinosporamide story—from a marine actinomycete-derived natural product to clinical trials,” we noted that the list is very short for marine natural products that have successfully progressed through advanced clinical trials or gained FDA approval for therapeutic use. This outcome is unusual because the idea of drugs from the sea has been on the table since the late 1960s [23]. There have been countless reviews that have examined the progress on this topic [24]. Nonetheless, some brief milestones include that in December 2004, the US Food and Drug Administration approved the first totally marine-derived drug, Prialt® to treat chronic and severe pain. This compound originally named ziconotide was isolated from the cone snail Conus magus [25,26,27]. By 2016, there were seven FDA or EMA approved small molecule drugs based on marine natural products [28], and as of 2020 the count is up to 12 [29]. Currently, there are many other promising scaffolds in the pipeline [29]. We believe that the most important current and future opportunities for marine inspired clinical therapeutics have and will continue to come from marine invertebrates (in particular sponges and tunicates) and bacteria. Examples of proof-of-concept outcomes will be discussed here, and the current successes illustrate that the past obstacles primarily associated with the supply problem during preclinical and clinical development can be overcome [30]. In this regard, the varying strategies that have been successful range from scale-up fermentation, total synthesis, invertebrate mariculture, and partial synthesis from precursors available from bacterial fermentation. As the exploration of the microbial origin of many invertebrate-derived natural products expands, strategies involving synthetic biology and the expression of key biosynthetic pathways are on the horizon. These roadblock busting strategies are often discussed in reviews and are briefly highlighted in some sections below.
The focus of our treatise next is to underscore the outstanding potential for drug development based on sponge, tunicate, and bacterial-derived natural products. Currently, 5 out of 12 marine-derived drugs in the FDA approved portfolio are from sponges (3) and tunicates (2) (Fig. 3). There are currently no approved therapeutics from marine bacteria, however, it is very important to note, as shown in Scheme 1, that a large percentage of sponge and tunicate overall mass is attributed to the associated microbiome of these organisms [31,32,33,34]. In the case of sponges, many have a significant microbial biomass (up to 35% of the total) and some of the most chemically rich taxa high in microbial abundance (HMA) sponges have been shown to have associations with both gram-positive and gram-negative bacteria. A similar situation can be seen for the tunicate holobiont—significant biomass of bacterial associates, dominated by gram-negative bacteria. We will discuss in greater detail below, the important role such bacteria play in the biosynthetic origin of these approved therapeutics.
A summary of therapeutics approved for clinical use or currently in Phase II/III clinical trials from sponges, tunicates, and bacteria is presented in Fig. 3. There are a larger number of marine natural products that have been examined in clinical trials and eventually dropped. Currently, there is no comprehensive review of such compounds, but important insights can be gleaned from a 2014 American Society of Pharmacognosy meeting lecture [35]. The assemblage of eight compounds in Fig. 3 includes representatives from all three of the taxa mentioned above. The tremendous developmental success for Sal A (2) with the preclinical work being driven by compound obtained from scale-up fermentation of the actinomycete Salinispora strain NPS21184 was discussed above (Fig. 2, section “The salinosporamide story—from a marine actinomycete-derived natural product to clinical trials”). The collection in Fig. 3 also includes the following sponge-derived (phylum Porifera) molecules. (1) Cytarabine (9) and vidarabine (10) are D-arabinose containing nucleosides that were isolated from Cryptotethya crypta and are considered by some as the first marine-derived approved pharmaceuticals [36]. Cytarabine is still used in treatments against various forms of leukemia and non-Hodgkin’s lymphoma [37], while vidarabine was an antiviral drug that is active against poxviruses, herpes viruses, some rhabdoviruses, hepadnaviruses, and RNA tumor viruses [38]. However, with the advancement of less toxic and more metabolically stable antivirals vidarabine was discontinued in the US in 2001 [29]. (2) Eribulin mesylate (11) (aka Halaven®) consists of the macrocyclic lactone pharmacophore of the natural product halichondrin B, a compound that was isolated in extremely low yields from Halichondria okadai and is hypothesized to be produced by an associated bacterium [39, 40]. This polyketide is currently used to treat metastatic breast cancer and inoperable liposarcoma [41]. Lastly, (3) plocabulin (12) was originally isolated from Lithoplocamia lithistoides and is currently in Phase II clinical trials for patients with advanced malignancies [42, 43].
A total of three tunicate-derived (phylum Chordata) natural products are in clinical use or late stage clinical trials. (1) ET-743 (13) (aka trabectedin, Yondelis®), isolated from Ecteinascidia turbinate, is currently in use to treat soft-tissue sarcoma and ovarian cancer [44]. (2) Lurbinectedin (14) (aka Zepsyre®) is a synthetic derivative of ET-743 that has been shown to have a substantially higher tolerated dose than its natural product counterpart [45], as well as higher overall survival, progression-free survival, and overall response rates in Phase III clinical trials against ovarian cancer [46]. (3) Plitidepsin (15) (aka Aplidin®, dehydrodidemnin B) was originally isolated from Aplidium albicans, has been approved in Australia for use against multiple myeloma [47], and has exhibited activity against the human coronavirus HCoV-229E, suggesting it could be an effective agent against the current outbreak of COVID-19 [48]. The bioactive 15, which was isolated from its tunicate host in high yields, is structurally related to didemnin B (67) from Tistrella bacteria [49, 50], and it is currently manufactured by total synthesis [51].
It is important to emphasize that during the discovery of some invertebrate-associated compounds, such as halichondrin B (sponge) and ET-743 (13) (tunicate), hypotheses were formulated and subsequently proven (see section “The possible bacterial biosynthetic origin of molecules isolated from sponges and tunicates—the case made by analyzing isolated yields, similar scaffolds, or results from culture-independent insights”) that the true biosynthetic sources for some molecules are associated bacteria. A summary of a few general supporting observations that stimulate additional inquiry on this topic is: (1) when similar classes of compounds are extracted from invertebrates of diverse taxonomic origin; (2) when a large percentage of some marine invertebrates’ overall mass is attributed to the associated microbiome (Scheme 1); and (3) when natural products isolated from invertebrates possess similar chemical scaffolds vs. that of bacterial natural products [52, 53].
The possible bacterial biosynthetic origin of molecules isolated from sponges and tunicates—the case made by analyzing isolated yields, similar scaffolds, or results from culture-independent insights
It is essential to further discuss four compounds shown above in Fig. 3 that are currently used as medicines and whose origins included sponges or tunicates. This list consists of: cytarabine (9), eribulin mesylate (11), ET-743 (13), and dehydrodidemnin B (15). Here is an important question: are their clues in the development trajectory of these compounds to imply that bacterial-derived pathways are functional in their biosynthesis? Another question—are these compounds present at extremely low concentrations from the invertebrate also implying a bacterial origin? Early on, Professor Fenical championed the bacterial origin theory stating in his 1993 review paper, “The importance of bacterial symbiosis is growing in recognition that bacteria may be the true producers of many compounds isolated from sponges, ascidians, and other marine invertebrates.” [54]. The possibility that true biosynthesis occurs by symbionts is directly relevant for two of the aforementioned compounds, 11 and 13. However, this does not apply to 9, which is isolated in high yields from sponges and discovered by medicinal chemistry. Also, the situation for 15 is perhaps enigmatic, as it was isolated in high yields from the Aplidium (tunicate) and is almost identical to didemnin B (67), which was obtained in high yields from Aplidium and in variable yields from marine-derived Tistrella bacteria [49, 50].
The design of eribulin mesylate (11) arose from the discovery and elucidation of halichondrin B (C60H86O19), a potent cytotoxin (IC50 = 0.09 ng/mL vs. B-16 melanoma cancer cells). Halichondrin B was isolated in miniscule yields from two disparate sponges, Halichondria okadai and Lissodendoryx sp. [39]. Its antitumor activity and pharmacophore were the inspiration for 11. Problematic for the preclinical development of halichondrin B was that a scant of 300 mg was obtained from 2000 pounds of Lissodendoryx, which thwarted its extensive preclinical follow-up. Currently, it is believed that dinoflagellate symbionts in the sponge are the true biosynthetic source of halichondrin B [55], and consistent with this idea is that the known dinoflagellate polyether okadaic acid was isolated from two species of Halichondria sponges [56].
A similar story to that sketched above is evident for ET-743 (13) first isolated in very low yields from the tunicate Ecteinascidia turbinate. Reports of anti-cellular proliferation were first described as early as 1969 when it was found that an extract of the Caribbean tunicate killed tumor cells in vitro and was capable of inhibiting tumor growth in vivo [57]. Due to the low abundance of this compound in the tunicate, the structure responsible for the described cytotoxic activity would not be published until 1990 and preclinical follow-up using natural material was not possible [58, 59].
Early on it was hypothesized that 13 and its congeners (16–18) shown in Fig. 4 were bacterial in origin as they shared similarities to chemical scaffolds of previously reported bacterial compounds including: saframycin A–C (19–21) from Streptomyces lavendulae [60, 61], saframycin Mx1 (22) from Myxococcus xanthus [62], and safracin A and B (23, 24) from Pseudomonas fluorescens [63]. In fact, safracin B (24) would later serve as the starting point in the semi-synthesis of Yondelis® (13) for clinical use [64]. Studies investigating the microbiome of E. turbinate in the Mediterranean [65] and Caribbean [66] found the γ-proteobacterium, Ca. E. frumentensis was the dominant member of the microbiome regardless of geographical location, furthering the bacterial origin hypothesis for 13. Then in 2011, the first confirmation of the bacterial origin of 13 was reported [67]. This study reported the assembly of a 35 kb contig from the tunicate metagenomic DNA consisting of 25 genes from the core of the non-ribosomal peptide synthase (NRPS) portion of the gene cluster associated with 13 production. During this work it was not possible to assemble the entire 13 gene cluster, yet the GC content and codon preference of the contig suggested that this NRPS belonged to Ca. E. frumentensis [67]. In 2015, 13 was concluded to be produced by the unculturable bacterial endosymbiont Ca. E. frumentensis, obtained directly from metagenomic DNA [2]. This study also identified that Ca. E. frumentensis has an extremely reduced genome, indicating it is in the later stages of symbiosis, and that direct fermentation to supply 13 may not be possible. However, scale-up production through semi-synthesis by PharmaMar in partnership with Johnson & Johnson has provided a sustainable source of the compound.
It is important to underscore several points contained in the preceding narrative. To date there have been no meaningful examples of mariculture as a cost-effective route for sponge or tunicate-derived compound scale-up [68]. Alternatively, total synthesis of complex chemical scaffolds possessing multiple chiral centers present in many bioactive sponge and tunicate natural products has been successful in moving compounds through preclinical evaluation (best example is discodermolide [69]). This was a strategy used to create eribulin mesylate (10), and as noted above semi-synthesis continues to be successful in providing ET-743 (13). Not to be forgotten is that there is much interest in the hypothesis that bacteria are the true producers of many bioactive compounds in marine invertebrates, and as understanding and tools continue to evolve there may be practical alternatives to provide scale-up production of these desired compounds [52, 53].
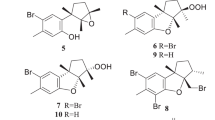
The advancement of next generation sequencing technology has allowed additional tunicate-derived natural products to be concluded to be bacterial in origin and ten examples are shown in Fig. 5. In 2005, the cytotoxic patellamides (25–27), originally isolated from Lissoclinum patella [70], were determined to be biosynthesised by the symbiotic cyanobacteria, Prochloron didemnid [71, 72]. Later, it was determined that the patellazoles (28–30), cytotoxic polyketides originally isolated from L. patella in 1988 [73, 74], were in fact biosynthesized by the symbiotic Candidatus Endolissoclinum faulkneri, a bacterium that belongs to the phylum Proteobacteria [75]. Lastly, in 2017, the biosynthetic gene cluster for the madelalides (31–34) was identified [76]. These mitochondrial ATP synthase inhibiting compounds, originally isolated from a Lissoclinum sp. [77,78,79], were determined to be produced from the symbiotic bacterium, Candidatus Didemnitutus mandela that belongs to the phylum Verrucomicrobia.
The cytotoxic onnamide A (35) and psymberin (36) shown in Fig. 6 were among the first sponge associated natural products to be identified as bacterial in origin through culture-independent methods [80,81,82,83,84]. The onnamides (35) were initially isolated from the sponge T. swinhoei and psymberin (36) from the sponges Psammocinia aff. bulbosa and Ircinia ramose [85,86,87]. However, the discovery of the biosynthetic origin for these compounds actually began with the structurally similar pederin and its initial isolation source, the terrestrial beetle Paederus fuscipes [88]. In 2004, the biosynthetic gene cluster for pederin was identified from the metagenome of P. fuscipes and determined that it originated from an uncultured Pseudomonas-like bacterium [89, 90]. Using the knowledge gained from the pederin biosynthetic gene cluster, researchers were able to probe the metagenome of the T. swinhoei and P. aff. bulbosa for homologous polyketide synthase domains. In 2004, they identified part of the biosynthetic gene cluster for onnamide A (35) from an unidentified bacterium in T. swinhoei [82], and in 2009 the nearly complete biosynthetic gene cluster for psymberin (36) was identified from an unknown bacterium in P. aff. bulbosa [84]. It was not until 2014, that the identity of the onnamide producing bacterium was determined to be the symbiont “Candidatus Entotheonella factor.” This bacterium would prove to be the source of many T. swinhoei compounds including the structurally similar theopederin A (37) [91]. However, mycalamide A (38) from a Mycale sponge while similar in structure to 35–37 from the very different sponge Theonella would be determined to originate from a completely different taxa of symbiont (to be discussed below) [92, 93]. Unfortunately, to date the identity of the psymberin producing bacterium is still unknown.
Briefly discussed above is that the bacterial genus Candidatus Entotheonella detected in sponges has proved to be a massive repository of biosynthetic richness, similar to what has been previously observed in soil actinomycetes (see section “Introduction”). Furthermore, these filamentous bacteria belong to the unique phylum Tectomicrobia and account for the majority of all known natural products isolated from T. swinhoei. [91, 94,95,96,97,98,99]. Distinct species of Candidatus Entotheonella have been shown to have symbiotic relationships with the different phenotypes of T. swinhoei (Y = yellow interior, W = white interior), perhaps accounting for the distinct chemotypes that have been previously observed in these sponges [100]. The Candidatus Entotheonella factor is the symbiont of T. swinhoei Y as shown in Fig. 7a and is responsible for the production of the onnamides (35) [85], theopederins (37) [101], polytheonamides (39, 40) [102], keramamides (41–43) [103], pseudotheonamides (44) [104], nazumamide A (45) [105], and the cyclotheonamides (46, 47) [106]. Representatives from each of these families of compounds are shown in Fig. 7a. The bacterial genus Candidatus Entotheonella serta is the symbiont of T. swinhoei W as shown in Fig. 7b and is responsible for the production of the swinholides (48–50) [107], misakinolides (51) [108], theopalauamide (52), and theonellamides (53–60) [109]. Representatives from each of these families of compounds are shown in Fig. 7b.
a Structures of Theonella swinhoei Y (phylum Porifera) natural products determined to originate from “Candidatus Entotheonella factor” through culture-independent methods. I = invertebrate source; B = bacterial producer. b Structures of Theonella swinhoei W (phylum Porifera) natural products determined to originate from “Candidatus Entotheonella serta” through culture-independent methods. I = invertebrate source; B = bacterial producer
Although the chemically prolific bacterial genus Candidatus Entotheonella, detected in T. swinhoei sponges, are the most well studied, it is also present in other sponge genera discussed in Fig. 8. In 2014, the metagenome of the sponge Discodermia calyx was mined for the biosynthetic gene cluster responsible for the production of the highly cytotoxic protein phosphatase inhibitor calyculin A (61) [110, 111]. This research identified the complete biosynthetic gene cluster and determined that it belonged to a bacterium of the genus Candidatus Entotheonella. However, the team discovered a phosphotransferase tailoring enzyme within the biosynthetic gene cluster suggesting a diphosphate compound may be the true end product of the biosynthetic pathway. Using a flash freeze-lyophilization extraction method on freshly collected D. calyx, they were able to isolate this new natural product, phosphocalyculin A (62). Interestingly, 62 exhibited a greater than 1000-fold reduction in cytotoxicity when compared with 61, suggesting that 62 is in fact the protoxin that D. calyx stores to avoid self-toxicity, but can quickly become the active agent in response to environmental stimuli [111]. The research discussed above highlights the powerful resource that metagenomic analysis can provide in the understanding of the chemical biology of sponge-derived natural products.
The preceding discussion illustrated that the bacterial genus Candidatus Entotheonella can be considered to be a “super-producer” of natural products. Culture-independent methods have also enabled the discovery of other sponge associated bacteria putatively responsible for the biosynthesis of complex natural products. As a striking example, in 2017, a metagenome exploration of Dysideidae sponges led to the identification of the biosynthetic gene clusters responsible for the production of cytotoxic polybrominated diphenyl ethers (PBDEs) (63, 64) [112]. This extended 2014 findings on the biosynthetic gene clusters responsible for the production of PBDEs from cultured γ-proteobacteria [113]. The next step was to close the loop by investigating the biosynthetic origin of PBDEs which are ubiquitous in Dysideidae sponges collected from coral reefs throughout the Indo-Pacific [114]. Interestingly, γ-proteobacteria were not concluded to be responsible for the production of PBDEs in Dysideidae, but instead originate from the sponge symbiotic cyanobacteria Hormoscilla spongeliae. Furthermore, the structural diversity of the PBDEs isolated from different Dysideidae samples appears correlated to the taxonomic clades of H. spongeliae strains [112].
The additional examples presented next, based on compounds shown in Figs. 6 and 8, further underscore that the biosynthetic production of complex sponge natural products can occur from various taxa of bacterial symbionts. One case involves mycalamide A (38) (Fig. 6) [115, 116], initially discovered in the 1990s from Mycale hentscheli. In 2020, two different groups independently exploring the metagenome [92, 93] of this sponge identified the biosynthetic pathway for 38 in the genome of Candidatus Entomycale ignis, a bacterium in the phylum Proteobacteria. Similarly, via additional metagenome guided research on M. hentscheli, the biosynthetic gene cluster for the translation initiation inhibitor pateamine A (65) (Fig. 8) [117, 118] was identified within the genome of Candidatus Patea custodiens, a bacterium in the phylum Kiritimatiellaeota. In addition, the biosynthetic gene cluster responsible for the production of the microtubule inhibitor peloruside A (66) [119, 120] was also identified within the metagenome of M. hentscheli. To date the taxonomy of the producing bacteria involved in the production of 66 has not been identified. Lastly, as part of the massive genetic diversity present in the metagenome of M. hentscheli, a polytheonamide-like ribosomally synthesized and posttranslationally modified peptide (similar in structure to 39 and 40) was identified within the genome of Candidatus Caria hoplite, a bacterium in the phylum Proteobacteria [93].
The preceding commentary dealing with bacterial biosynthetic gene clusters from invertebrate metagenomes, especially sponge metagenomes, emphasizes the existence of an immense range of natural product genetic diversity. It is clear that future metagenomic-driven research on invertebrates, such as those producing compounds shown in Figs. 4–8, will further reveal undiscovered natural products possessing inspirational structures.
The possible bacterial biosynthetic origin of molecules isolated from sponges and tunicates—the case made by analyzing results of cultured biosynthetic production
In the section above, we discussed the structures of more than 60 complex molecules directly isolated from sponges and tunicates. The focus was on assessing their chemodiversity alongside insights of the bacterial-derived biosynthetic machinery involved in their creation. In the future, synthetic biology coupled with the functional biosynthetic gene clusters identified in the metagenomes could provide a supply of important natural products [121]. A current roadblock is that in many cases the bacteria producing the natural products are obligate symbionts and are incapable of independent growth outside of their invertebrate host [122]. It is useful to explore situations where molecules analogous to those produced by sponges and tunicates can be successfully obtained by direct bacterial culturing in parallel with actual compound isolation. We suggest that insights obtained from such a survey could provide answers to the vexing question—what would it take to find new natural product chemical space? On the one hand, it is relevant to note that bacterial genome sequencing sometimes reveals that an individual strain can contain the machinery to produce more than 20 distinct molecules. This has stimulated many groups to contend that molecules encoded within the genome of bacterial isolates that are not obtained from culturing persist as “cryptic” entities whose pathways need to be turned-on [123, 124].
The situation with didemnin B (67) (Fig. 9a) provides an important case example. This compound was originally isolated in 1978 from a Trididemnum tunicate and exhibits a wide range of biological activity [125]. To date, nine congeners (didemnins A–E, G, X, Y, and nordidemnin B) have been isolated from extracts of Trididemnum solidum, and 67 possesses the most potent biological activities [126]. Relatively large amounts of didemnin B were isolated from the tunicate and even larger amounts were obtained by total synthesis. Unfortunately, high toxicity during Phase II clinical trials led to termination of further trials on 67. However, as discussed in Fig. 3 the analogue dehydrodidemnin B (15) successfully progressed through clinical trials and is now approved for therapeutic use. Especially important for this discussion was the serendipitous isolation of 67 through laboratory culture of five strains of Tistrella four mobilis, and one bauzanensis (see Fig. 9a) [49, 50]. The unoptimized compound yields from the culturing of two T. mobilis strains YIT 12409 and KA081020-065 were modest and included 67 (YIT 12409 = 3.2 mg/L; KA081020-065 = 0.2 mg/L) as well as nordidemnin B (YIT 12409 = 0.5 mg/L; KA081020-065 = 0.1 mg/mL). A detailed annotation of the biosynthetic gene cluster for 67 is in hand for Tistrella sp. [50] and suggests that synthetic biology coupled with semi-synthesis could provide an alternative scale-up route to 67, which for now is obtained by total synthesis.
a Structures of tunicate (phylum Chordata) and sponge (phylum Porifera) natural products determined to be bacterial in origin through culture-dependent methods. I = invertebrate source; B = bacterial producer. b Structures of sponge (phylum Porifera) natural products determined to be bacterial in origin through culture-dependent methods. I = invertebrate source; B = bacterial producer
There are four additional cases involving tunicate-associated natural products that have been isolated from cultured bacteria, and these are listed in Fig. 9a. For each, a member of the compound family was identified from their bacterial source prior to or simultaneously to being observed from invertebrate source. Initially, lissoclinolide (68) exhibiting antibiotic and antitumor activity was obtained from the terrestrial Actinobacteria, Micropolyspora venezuelensis in 1969, then >20 years later 68 was re-isolated from the tunicate Lissoclinum patella [127,128,129]. In 1976, the antibiotic enterocin (69) was identified from a terrestrial soil Streptomyces and in 1996 there are two reports of 69 being isolated, one from a marine Streptomyces maritimus and the other from the tunicate Didemnum [130,131,132]. Staurosporine (70) is a ubiquitous alkaloid commonly isolated from various terrestrial and marine Streptomyces and in 1992 was obtained from the tunicate Eudistoma toealensis [133, 134]. Lastly, the cytotoxic haterumalides was simultaneously isolated in 1999 from three disparate sources: (1) haterumalide NA (71) from Serratia marcescens, a terrestrial Proteobacteria, (2) 71 along with congeners haterumalides NB-NE (72–75) from an Ircinia sponge, and (3) 76 from a Lissoclinum tunicate [135,136,137].
There are a few cases where cultured bacterial isolates contained compounds also isolated from sponges. Two of these were discussed above and include nazumamide A (45) and PBDEs (63, 64) (Figs. 7a and 8). The situation for 45 is somewhat unique, this T. swinhoei sponge-derived compound was identified by culture-independent methods from in the genome of the T. swinhoei symbiont, Candidatus Entotheonella [91, 105] and also from the culturing of the actinomycete Salinispora pacifica [138]. The biosynthetic machinery for the Dysideidae sponge-derived 63, 64 was identified within the genome of cyanobacterial symbionts H. spongeliae, in addition to PBDE’s isolated from other cultured γ-proteobacteria [112, 113].
An additional 13 examples of sponge-derived compounds also isolated from cultured bacteria, shown in Fig. 9b, are discussed next. Toyocamycin (77) an antibiotic nucleoside was first isolated in 1965 from a terrestrial Streptomyces [139] and then decades later from the sponge Jaspis johnstoni [140]. Bacteriohopanetetrol (78) has been isolated from various taxa of bacteria, but its initial isolation was from the Proteobacteria Acetobacter xylinum in 1976 [141] and in 2001 it was identified in high yields in the sponge Plakortis simplex [142]. Manzamine A (79) exhibits a wide range of biological activity and since 1986 has been isolated in high yields from multiple taxa of sponges [143]. Later, it was isolated in miniscule and irreproducible yields from an actinobacteria Micromonospora strain [144]. The cytotoxic heterocycles bengamide E (80) and E′ (81), initially isolated from a Jaspis sponge [145], were later obtained in high and reproducible yields by culturing the terrestrial Myxococcus virescens [146]. The antifungal microsclerodermins were initially isolated from Microscleroderma and Theonella sponges [147, 148] and in 2013 microsclerodermin D (82) and L (83) were isolated from cultured Sorangium cellulosum and a Jahnella sp., bacteria belonging to the phylum Proteobacteria [149]. Lastly, a series of bromotyrosine-derived alkaloids isolated from various sponge sources were obtained but not always reproducibly from the Proteobacteria, Pseudovibrio denitrificans [150]. This included the cytotoxic fistularin-3 (84) [151], aerothionin (85), hydroxyaerothionin (86) [152], aplysinamisine II (87) [153], purealidin L (88) [154], and homopurpuroceratic acid (89) [155].
The possible bacterial biosynthetic origin of molecules isolated from sponges and tunicates—the case made by analyzing of biosynthetic production from both culture-dependent and independent schemes
Harnessing the power of metagenomics, synthetic biology, and culturable isolates potentially provides a route to unlock access to diverse chemical scaffolds often only seen in the metagenomes of invertebrates. The following two cases based on the compounds shown in Fig. 10 highlight different strategies in which these tools can assist in the discovery of invertebrate-associated natural products.
The anti-HIV peptide divamide A (90) was isolated from the ubiquitous tunicate, Didemnum molle, however, only nanogram quantities were obtained hindering structural elucidation. Metagenomic sequencing was used to predict structural features and the symbiotic cyanobacteria Prochloron didemnid was identified as the biosynthetic source of this peptide. A proof-of-concept result was provided by expressing the divamide pathway in E. coli which provided better quantities of divamide A (90) and of 11 other congeners. This latter outcome facilitated structural elucidation and biological screening [156].
Lobatamide A (91) was initially isolated from the tunicate Aplidium lobatum in 1998. Over two decades later, genome mining focused on identifying the products of diverse oxidation and directed researchers to focus on the polyketide synthases-associated flavoprotein monooxygenases involved in the biosynthesis of oxygen containing polyketides. The biosynthetic modules identified from the plant-derived Proteobacteria, Gynuella sunshinyii [157], were identified as responsible for the oxygen incorporation in the biosynthesis of 91, and also provided a biosynthetic hypothesis for insertion of oxygen atoms into the macrocyclic cores of sponge-derived salarin A and pateamine A (65). A 5 L culture provided an unspecified amount of 91 that was rigorously characterized by NMR and MS.
Prospects to gather future understanding on the involvement of bacteria in producing meaningful metabolites from sponges and tunicates
Scheme 1 and the accompanying annotations highlight bacterial diversity associated with the holobiont of sponges and tunicates. Also, the remarks in Figs. 3–10 summarize bacterial taxa possessing prolific machinery for natural product biosynthesis. Prior to the advancements and general availability of next generation sequencing it was often hypothesized, without buttressing experimental data, that sponge and tunicate-associated natural products were bacterial in origin especially when similar chemical scaffolds were isolated from both sources. This section focuses on three sets of our favorite sponge natural products that have similar scaffolds to the bacterial compounds as shown in Fig. 11. Most importantly, current knowledge is incomplete about the biosynthetic synergy between the invertebrates and bacteria that produce this collection of compounds. First, jasplakinolide (92) and its >20 congeners have been isolated from at least three different taxa of sponges [158, 159]. This family of compounds contain structural similarities to the chondramides (93–96) produced by the terrestrial bacteria Chondromyces crocatus, and the miuraenamides (97–99) produced by the marine bacteria Paraliomyxa miuraensis [160, 161]. Second, salicylihalamide A (100) isolated from the Haliclona sponge [162] is structurally similar to apicularen A (101) produced by the terrestrial bacteria, Chondromyces robustus [163]. Last, latrunculin A (102), initially isolated from the sponge Latrunculia magnifica [164], shares structural similarities with the epothilones (103, 104) produced by the terrestrial bacteria, Sorangium cellulosum [165].
Conclusions
An overarching goal in this review has been to illustrate the evolving overlap between the biosynthetic machinery of marine-derived bacteria vs. that of the chemically prolific sponges and tunicates. The ~28,500 marine natural products identified by the end of 2016 constitute an enormous assemblage of wide-ranging structural scaffolds. Annually, more than 1000 new compounds are described from Oceania and in 2018 the annual total tally was 1554 compounds: 222 from sponges, 12 from tunicates, and 240 from marine bacteria (with 69% of these from actinomycetes) [81]. A striking theme contained in both recent research papers and reviews outlines that several sponge/tunicate-derived bioactive compounds are seemingly produced by the action of the invertebrate microbiome.
Discussed in this review were examples describing biosynthetic outcomes producing an immense range of natural product structural diversity, potentially arising through synergy between gene clusters from sponge, tunicate, and bacteria metagenomes. At the top of the list of examples in the review are: (1) the molecular genetics-based discoveries from the sponge T. swinhoei, rich with the unculturable symbiont Candidatus Entotheonella, and (2) the use of complete genome sequences from the actinomycete S. tropica to direct further molecule discovery. These and many other case examples presented in this review dramatically illustrate the potential for selected sponges, tunicates, and marine-derived bacteria to provide an inexhaustible supply of novel natural products. Needed at this juncture is a firm understanding of the true nature of sponge/tunicate-microorganism symbiont interactions whose machineries produce novel metabolites in the natural environment that could potentially be followed-up by carrying out the natural product production in the laboratory. Such new understanding will undoubtedly reveal fresh paradigms in marine natural products research and this is underscored by the dramatic statement in a 2020 review, “We continue to draw the attention of readers to the recognition that a significant number of natural product drugs/leads are actually produced by microbes and/or microbial interactions with the host from whence it was isolated” [24]. We predict that in the future new insights will be obtained by focusing on parallel scaffolds from sponges and tunicates vs. those of marine-derived bacteria. The toolbox for the future to discover new paradigms in marine natural products research must combine analytical spectrometry with the application of synthetic biology, genome mining, experimental therapeutics, and other transformative approaches.
References
Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2009;26:338–62.
Schofield MM, Jain S, Porat D, Dick GJ, Sherman DH. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ Microbiol. 2015;17:3964–75.
Lackner G, Peters EE, Helfrich EJ, Piel J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc Nat Acad Sci. 2017;114:E347–56.
Roach AW, Silvey JKG. The occurrence of marine actinomycetes in Texas Gulf Coast substrates. Am Midl Nat. 1959;62:482–99.
Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol. 2002;68:5005–11.
Moore BS, Gerwick WH. Special issue in honor of William Fenical, a pioneer in marine natural products discovery and drug development. J Nat Prod. 2015;78:347–8.
Renner MK, Shen YC, Cheng XC, Jensen PR, Frankmoelle W, Kauffman CA, et al. New antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). J Am Chem Soc. 1999;121:353–60.
Jensen PR, Moore BS, Fenical W. The marine actinomycete genus Salinispora: a model organism for secondary metabolite discovery. Nat Prod Rep. 2015;32:738–51.
Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Sporolides A and B: structurally unprecedented halogenated macrolides from the marine actinomycete Salinispora tropica. Org Lett. 2005;7:2731–4.
Hughes CC, Prieto-Davo A, Jensen PR, Fenical W. The marinopyrroles, antibiotics of an unprecedented structure class from marine Streptomyces sp. Org Lett. 2008;10:629–31.
Hughes CC, MacMillan JB, Gaudêncio SP, Jensen PR, Fenical W. The ammosamides: structures of cell cycle modulators from a marine-derived streptomyces species. Angew Chem Int Ed. 2009;48:725–7.
Ting CP, Funk MA, Halaby SL, Zhang Z, Gonen T, van der Donk WA. Use of a scaffold peptide in the biosynthesis of amino acid–derived natural products. Science. 2019;365:280–4.
Kaysser L, Bernhardt P, Nam SJ, Loesgen S, Ruby JG, Skewes-Cox P, et al. Merochlorins A-D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. J Am Chem Soc. 2012;134:11988–91.
Brandstätter M, Freis M, Huwyler N, Carreira EM. Total synthesis of (−)-merochlorin A. Angew Chem Int Ed. 2019;58:2490–4.
Jang KH, Nam SJ, Locke JB, Kauffman CA, Beatty DS, Paul LA, Fenical W. Anthracimycin, a potent anthrax antibiotic from a marine-derived actinomycete. Angew Chem Int Ed. 2013;52:7822–4.
Kim MC, Machado H, Jang KH, Trzoss L, Jensen PR, Fenical W. Integration of genomic data with nmr analysis enables assignment of the full stereostructure of neaumycin B, a potent inhibitor of glioblastoma from a marine-derived micromonospora. J Am Chem Soc. 2018;140:10775–84.
US National Institutes of Health. A phase III trial of marizomib in patients with newly diagnosed glioblastoma (NCT03345095). https://clinicaltrials.gov/ct2/show/NCT03345095. Accessed 3 May 2020.
Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg Med Chem. 2009;17:2175–80.
Potts BC, Lam KS. Generating a generation of proteasome inhibitors: from microbial fermentation to total synthesis of salinosporamide A (Marizomib) and other salinosporamides. Mar Drugs. 2010;8:835–80.
Lam KS, Kenneth L, Neuteboom STC, Palladino MA, Sethna KM, Spear MA, et al. From natural product to clinical trials: NPI-0052 (salinosporamide A), a marine actinomycete-derived anticancer agent. In: Buss AD, Butler MS, editors. Natural product chemistry for drug discovery. Cambridge: Royal Society of Chemistry Publishing; 2009: p. 355–73.
Corey EJ, Li WD. Total synthesis and biological activity of lactacystin, omuralide and analogs. Chem Pharm Bull. 1999;47:1–10.
Gholami H, Kulshrestha A, Favor OK, Staples RJ, Borhan B. Total synthesis of (−)‐salinosporamide A via a late stage C− H insertion. Angew Chem Int Ed. 2019;58:10110–3.
Freudenthal HD, editor. In: Drugs from the sea: transactions of the drugs from the sea symposium University of Rhode Island. Washington, DC: Marine Technology Society; 1968.
Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;27:770–803.
McIntosh M, Cruz LJ, Hunkapillar MW, Gray WR, Olivera BM. Isolation and structure of a peptide toxic from the marine snail Conus magus. Arch Biochem Biophys. 1982;218:329–34.
McGivern JG. Ziconotide: A review of its pharmacology and use in the treatment of pain. Neuropsychiatr Dis Treat. 2007;3:69–85.
Brookes ME, Eldabe S, Batterham A. Ziconotide monotherapy: a systematic review of randomised controlled trials. Curr Neuropharmacol. 2017;15:217–31.
Lindequist U. Marine-derived pharmaceuticals—challenges and opportunities. Biomol Ther. 2016;24:561–71.
Clinical Pipeline Marine Pharmacology. www.midwestern.edu/departments/marinepharmacology/clinical-pipeline.xml. Accessed 3 May 2020.
Proksch P, Edrada-Ebel R, Ebel R. Drugs from the sea—opportunities and obstacles. Mar Drugs. 2003;1:5–17.
Gloeckner V, Wehrl M, Moitinho-Silvia L, Gernert C, Schupp P, Pawlik JR, et al. The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol Bull. 2014;227:78–88.
Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, et al. Diversity, Structure and convergent evolution of the global sponge microbiome. Nat Commun. 2016;7:11870.
Moitinho-Silva L, Steinert G, Nielsen S, Hardoim CCP, Wu YC, McCormack GP, et al. Predicting the HMA-LMA status is marine sponges by machine learning. Front Microbiol. 2017;8:752.
Pita L, Rix L, Slaby BM, Franke A, Hentschel U. The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome. 2018;6:46.
Cragg GM, Newman DJN. Natural products. A history of success and continuing promise for drug discovery and development. http://www.pharmacognosy.us/wp-content/uploads/ASP-Presentation-2014.pdf. Accessed 14 May 2020.
Bergmann W, Burke DC. Contributions to the study of marine products. XXXIX. The nucleosides of sponges. III. Spongothymidine and spongouridine. J Org Chem. 1955;20:1501–7.
Lichtman MA. A historical perspective on the development of the cytarabine (7 days) on daunorubicin (3 days) trearment regimen for acute myelogenous leukemia: 2013 the 40th anniversary of 7+3. Blood Cells Mol Dis. 2013;50:119–30.
Shen W, Kim JS, Kish PE, Zhang J, Mitchell S, Gentry BG, et al. Design and synthesis of vidarabine prodrugs as antiviral agents. Bioorg Med Chem Lett. 2009;19:792–6.
Hirata Y, Uemura D. Halichondrins—antitumor polyether macrolides from a marine sponge. Pure Appl Chem. 1986;58:701–10.
Towle MJ, Salvato KA, Budrow J, Wels BF, Kuznetsov G, Aalfs KK, et al. In vitro and in vivo anticancer activities of synthetic macrocyclic ketone analogues of halichondrin B. Cancer Res. 2001;61:1013–21.
Aseyev O, Ribeiro JM, Cardoso F. Review on the clinical use of eribulin mesylate for the treatment of breast cancer. Expert Opin Pharmacother. 2016;17:589–600.
Pera B, Barasoain I, Pantazopoulou A, Canales A, Matesanz R, Rodriguez-Salarichs J, et al. New interfacial microtubule inhibitors of marine origin, PM050489/PM060184, with potent antitumor activity and a distinct mechanism. ACS Chem Biol. 2013;8:2084–94.
Galmarini CM, Martin M, Bouchet BP, Guillen-Navarro MJ, Martínez-Diez M, Martinez-Leal JF, et al. Plocabulin, a novel tubulin-binding agent, inhibits angiogenesis by modulation of microtubule dynamics in endothelial cells. BMC Cancer. 2018;18:164.
Gordon EM, Sankhala KK, Chawla N, Chawla SP. Trabectedin for soft tissue sarcoma: current status and future perspectives. Adv Ther. 2016;33:1055–71.
Elez ME, Tabernero J, Geary D, Macarulla T, Kang SP, Kahatt C, et al. First-in-human phase I study of lurbinectedin (PM01183) in patients with advanced solid tumors. Clin Cancer Res. 2014;20:2205–14.
Gaillard S, Ghamande SA, Pardo B, Lorusso D, Vergote A, Papai Z, et al. CORAIL trial: randomized phase III study of lurbinectedin (PM01183) versus pegylated liposomal doxorubicin (PLD) or topotecan (T) in patients with platinum-resistant ovarian cancer. J Clin Oncol. 2016;34:15.
Delgado-Calle J, Kurihara N, Atkinson EG, Nelson J, Miyagawa K, Galmarini CM, et al. Aplidin (Plitidepsin) is a novel anti-myeloma agent with potent anti-resorptive activity mediated by direct effects on osteoclasts. Oncotarget. 2019;10:2709–21.
BioSpace. PharmaMar has announced that the Spanish medicines agency has authorized the APLICOV-PC clinical trial with Aplidin® (Plitidepsin) for the treatment of patients with COVID-19. 2020. www.biospace.com/article/releases/pharmamar-has-announced-that-the-spanish-medicines-agency-has-authorized-the-aplicov-pc-clinical-trial-with-aplidin-plitidepsin-for-the-treatment-of-patients-with-covid-19. Accesses 4 May 2020.
Tsukimoto M, Nagaoka M, Shishido Y, Fujimoto J, Nishisaka F, Matsumoto S, et al. Bacterial production of the tunicate-derived antitumor cyclic depsipeptide didemnin B. J Nat Prod. 2011;74:2329–31.
Xu Y, Kersten RD, Nam SJ, Lu L, Al-Suwailem AM, Zheng H, et al. Bacterial biosynthesis and maturation of the didemnin anti-cancer agents. J Am Chem Soc. 2012;134:8625–32.
Alonso-Álvarez S, Pardal E, Sánchez-Nieto D, Navarro M, Caballero MD, Mateos MV, et al. Plitidepsin: design, development, and potential place in therapy. Drug Des Devel Ther. 2017;11:253–64.
Gerwick WH, Moore BS. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem Biol. 2012;19:85–98.
Still PC, Johnson TA, Theodore CM, Loveridge ST, Crews P. Scrutinizing the scaffolds of marine biosynthetics from different source organisms: gram-negative cultured bacterial products enter center stage. J Nat Prod. 2014;77:690–702.
Fenical W. Chemical studies of marine bacteria: developing a new resource. Chem Rev. 1993;93:1673–83.
Van Wagoner RM, Satake M, Wright JL. Polyketide biosynthesis in dinoflagellates: what makes it different? Nat Prod Rep. 2014;31:1101–37.
Tachibana K, Scheuer PJ, Tsukitani Y, Kikuchi H, Van Engen D, Clardy J, et al. Okadaic acid, a cytotoxic polyether from two marine sponges of the genus Halichondria. J Am Chem Soc. 1981;103:2469–71.
Sigel MM, Wellham LL, Lichter W, Dudeck LE, Gargus JL, Lucas LH. In: Youngken HW, editor. Food-drugs from the sea: proceedings 1969. Washington DC: Marine Technology Society; 1970. p. 281–94.
Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, et al. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: potent antitumor agents from the caribbean tunicate Ecteinascidia turbinata. J Org Chem. 1990;55:4512–5.
Wright AE, Forleo DA, Gunawardana GP, Gunasekera SP, Koehn FE, McConnell OJ. Antitumor tetrahydroisoquinoline alkaloids from the colonial ascidian Ecteinascidia turbinata. J Org Chem. 1990;55:4508–12.
Arai T, Takahashi K, Kubo A. New antibiotics saframycins A, B, C, D and E. J Antibiot. 1977;30:1015–8.
Arai T, Takahashi K, Kubo A, Nakahara S, Sato S, Aiba K, et al. The structures of novel antibiotics, saframycin B and C. Tetrahedron Lett. 1979;20:2355–8.
Irschik H, Trowitzsch-Kienast W, Gerth K, Hofle G, Reichenbach H. Saframycin Mx1, a new natural saframycin isolated from a myxobacterium. J Antibiot. 1988;41:993–8.
Ikeda Y, Idemoto H, Hirayama F, Yamamoto K, Iwao K, Asao T, et al. Safracins, new antitumor antibiotics. I. Producing organism, fermentation and isolation. J Antibiot. 1983;36:1279–83.
Cuevas C, Francesch A. Development of Yondelis (Trabectedin, ET-743). A semisynthetic process solves the supply problem. Nat Prod Rep. 2009;26:322–37.
Moss C, Green DH, Pérez B, Velasco A, Henríquez R, McKenzie JD. Intracellular bacteria associated with the ascidian Ecteinascidia turbinata: phylogenetic and in situ hybridisation analysis. Mar Biol. 2003;143:99–110.
Pérez-Matos AE, Rosado W, Govid NS. Bacterial diversity associated with the caribbean tunicate Ecteinascidia turbinate. Antonie Van Leeuwenhoek. 2007;92:155–64.
Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem Biol. 2011;6:1244–56.
Schaufelberger DE, Koleck MP, Beutler JA, Vatakis AM, Alvarado AB, Andrews P, et al. The large-scale isolation of bryostatin 1 from bugula neritina following current good manufacturing practices. J Nat Prod. 1991;54:1265–70.
Smith AB III, Oiu Y, Jones DR, Kobayashi K. Total synthesis of (-)-discodermolide. J Am Chem Soc. 1995;117:12011–2.
Ireland CM, Durso AR Jr, Newman RA, Hacker MP. Antineoplastic cyclic peptides from the marine tunicate Lissoclinum patella. J Org Chem. 1982;47:1807–11.
Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci. 2005;102:7315–20.
Long PF, Dunlap WC, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. ChemBioChem. 2005;6:1760–5.
Corley DG, Moore RE, Paul VJ. Patellazole B: a novel cytotoxic thiazole-containing macrolide from the marine tunicate Lissoclinum patella. J Am Chem Soc. 1988;110:7920–2.
Phillips AW, Anketell MJ, Balan T, Lam NYS, Williams S, Paterson I. Toward the total synthesis of patellazole B: synthesis of an advanced C1-C25 fragment corresponding to the macrocyclic skeleton. Org Biomol Chem. 2018;16:8286–91.
Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Nat Acad Sci. 2012;109:20655–60.
Lopera J, Miller IJ, McPhail KL, Kwan JC. Increased biosynthetic gene dosage in a genome-reduced defensive bacterial symbiont. mSystems. 2017;2:e00096–17.
Sikorska J, Hau AM, Anklin C, Parker-Nance S, Davies-Coleman MT, Ishmael JE, et al. Mandelalides A–D, cytotoxic macrolides from a new lissoclinum species of South African tunicate. J Org Chem. 2012;77:6066–75.
Nazari M, Serrill JD, Sikorska J, Ye T, Ishmael JE, McPhail KL. Discovery of mandelalide E and determinants of cytotoxicity for the mandelalide series. Org Lett. 2016;18:1374–7.
Nazari M, Serrill JD, Wan X, Nguyen MH, Anklin C, Gallegos DA, et al. New mandelalides expand a macrolide series of mitochondrial inhibitors. J Med Chem. 2017;60:7850–62.
Blunt JW, Copp BR, Keyzers RA, Munro MHG, Prinsep MR. Marine natural products. Nat Prod Rep. 2017;17:235–94.
Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep. 2020;37:175–223.
Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci. 2004;101:16222–7.
Piel J, Hui D, Fusetani N, Matsunaga S. Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ Microbiol. 2004;6:921–7.
Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat Chem Biol. 2009;5:494–501.
Sakemi S, Ichiba T, Kohmoto S, Saucy G, Higa T. Isolation and structure elucidation of onnamide A, a new bioactive metabolite of a marine sponge, Theonella sp. J Am Chem Soc. 1988;110:4851–3.
Cichewicz RH, Valeriote FA, Crews P. Psymberin, a potent sponge-derived cytotoxin from Psammocinia distantly related to the pederin family. Org Lett. 2004;6:1951–4.
Pettit GR, Xu JP, Chapuis JC, Pettit RK, Tackett LP, Doubek DL, et al. Antineoplastic Agents. 520. Isolation and structure of ircinia statins A and B from the Indo-Pacific marine sponge Ircinia ramosa. J Med Chem. 2004;47:1149–52.
Cardani C, Fuganti C, Ghiringhelli D, Grasselli P, Pavan M, Valcurone MD. The biosynthesis of pederin. Tetrahedron Lett. 1973;30:2815–8.
Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of paederus beetles. Proc Natl Acad Sci. 2002;99:14002–7.
Piel J, Wen G, Platzer M, Hui D. Unprecedented diversity of catalytic domains in the first four modules of the putative pederin polyketide synthase. ChemBioChem. 2004;5:93–8.
Wilson MC, Mori T, Rückert C, Uria AR, Helf MJ, Takada K, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62.
Rust M, Helfrich EJN, Freeman MF, Nanudorn P, Field CM, Rückert C, et al. A multiproducer microbiome generates chemical diversity in the marine sponge Mycale hentscheli. Proc Natl Acad Sci. 2020;117:9508–18.
Storey MA, Andreassend SK, Bracegirdle J, Brown A, Keyzers RA, Ackerley DF, et al. Metagenomic exploration of the marine sponge Mycale hentscheli uncovers multiple polyketide-producing bacterial symbionts. mBio. 2020;11:e02997–19.
Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis”. Mar Biol. 2000;136:969–77.
Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, et al. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338:387–90.
Ueoka R, Uria AR, Reiter S, Mori T, Karbaum P, Peters EE, et al. Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. Nat Chem Biol. 2015;11:705–12.
Smith DRM, Uria AR, Helfrich EJN, Milbredt D, van Pee KH, Piel J, et al. An unusual flavin-dependent halogenase from the metagenome of the marine sponge Theonella swinhoei WA. ACS Chem Biol. 2017;12:1281–7.
Freeman MF, Helf MJ, Bhushan A, Morinaka BI, Piel J. Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium. Nat Chem. 2017;9:387–95.
Mori T, Cahn JKB, Wilson MC, Meoded RA, Wiebach V, Martinez AFC, et al. Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated entotheonella sponge symbionts. Proc Natl Acad Sci. 2018;115:1718–23.
Wegerski CJ, Hammond J, Tenney K, Matainaho T, Crews P. A serendipitous discovery of isomotuporin-containing sponge populations of Theonella swinhoei. J Nat Prod. 2007;70:89–94.
Fusetani N, Sugawara T, Matsunaga S. Bioactive marine metabolites. 41. Theopederins A-E, potent antitumor metabolites from a marine sponge, Theonella sp. J Org Chem. 1992;57:3828–32.
Hamada T, Matsunaga S, Yano G, Fusetani N. Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge, Theonella swinhoei. J Am Chem Soc. 2005;127:110–8.
Kobayashi J, Sato M, Ishibashi M, Shigemori H, Nakamura T, Ohizumi Y. Keramamide A, a novel peptide from the okinawan marine sponge Theonella sp. J Chem Soc Perkin Trans. 1991;1:2609–11.
Nakao Y, Masuda A, Matsunaga S, Fusetani N. Pseudotheonamides, serine protease inhibitors from the marine sponge Theonella swinhoei. J Am Chem Soc. 1999;121:2425–31.
Fusetani N, Nakao Y, Matsunaga S. Nazumamide A, a thrombin-inhibitory tetrapeptide, from a marine sponge, Theonella sp. Tetrahedron Lett. 1991;32:7073–4.
Fusetani N, Matsunaga S, Matsumoto H, Takebayashi Y. Bioactive marine metabolites. 33. Cyclotheonamides, potent thrombin inhibitors, from a marine sponge Theonella sp. J Am Chem Soc. 1990;112:7053–4.
Carmely S, Kashman Y. Structure of swinholide-A, a new macrolide from the marine sponge Theonella swinhoei. Tetrahedron Lett. 1985;26:511–4.
Sakai R, Higa T, Kashman Y. Misakinolide-A, an antitumor macrolide from the marine sponge Theonella sp. Chem Lett. 1986;15:1499–502.
Matsunaga S, Fusetani N. Theonellamides A-E, cytotoxic bicyclic peptides, from a marine sponge Theonella sp. J Org Chem. 1995;60:1177–81.
Kato Y, Fusetani N, Matsunaga S, Hashimoto K, Fujita S, Furuya T. Bioactive marine metabolites. Part 16. Calyculin A. A novel antitumor metabolite from the marine sponge Discodermia calyx. J Am Chem Soc. 1986;108:2780–1.
Wakimoto T, Egami Y, Nakashima Y, Wakimoto Y, Mori T, Awakawa T, et al. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat Chem Biol. 2014;10:648–55.
Agarwal V, Blanton JM, Podell S, Taton A, Schorn MA, Busch J, et al. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat Chem Bio. 2017;13:537–43.
Agarwal V, El Gamal AA, Yamanaka K, Poth D, Kersten RD, Schorn M, et al. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Biol. 2014;10:640–7.
Calcul L, Chow R, Oliver AG, Tenney K, White KN, Wood AW, et al. NMR strategy for unraveling structures of bioactive sponge-derived oxy-polyhalogenated diphenyl ethers. J Nat Prod. 2009;72:443–9.
Perry NB, Blunt JW, Munro MHG. Antiviral and antitumor agents from a New Zealand sponge, Mycale sp. 2. Structures and solution conformations of mycalamides A and B. J Org Chem. 1990;55:223–7.
Dyshlovoy SA, Fedorov SN, Kalinovsky AI, Shubina LK, Bokemeyer C, Stonik VA, et al. Mycalamide A shows cytotoxic properties and prevents egf-induced neoplastic transformation through inhibition of nuclear factors. Mar Drugs. 2012;10:1212–24.
Northcote PT, Blunt JW, Munro MHG. Pateamine: a potent cytotoxin from the New Zealand marine sponge Mycale sp. Tetrahedron Lett. 1991;32:6411–4.
Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, et al. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell. 2005;20:709–22.
West LM, Northcote PT, Battershill CN. Peloruside A: a potent cytotoxic macrolide isolated from the New Zealand marine sponge Mycale sp. J Org Chem. 2000;65:445–9.
Hood KA, West LM, Rouwé B, Northcote PT, Berridge MV, Wakefield SJ, et al. Peloruside A, a novel antimitotic agent with paclitaxel-like microtubule-stabilizing activity. Cancer Res. 2002;62:3356–60.
Huo L, Hug JJ, Fu C, Bian X, Zhang Y, Müller R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat Prod Rep. 2019;36:1412–36.
Kwan JC. The who, why, and how of small-molecule production in invertebrate microbiomes: basic insights fueling drug discovery. mSystems. 2018;3:e00186–17.
Udwary DW, Zeigler L, Asolker RN, Singan V, Lapidus A, Fenical W, et al. Genome sequencing reveals complex secondary metabolome in marine actinomycete Salinispora tropica. Proc Nat Acad Sci. 2007;104:10376–81.
Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J Am Chem Soc. 2010;132:2469–93.
Rinehart KL, Gloer JB, Cook JC Jr, Mizsak SA, Scahill TA. Structures of the didemnins, antiviral and cytotoxic depsipeptides from the Caribbean tunicate. J Am Chem Soc. 1981;103:1857–9.
Rinehart KL, Kishore V, Bible KC, Sakai R, Sullins DW, Li KM. Didemnins and tunichlorin: novel natural products from the marine tunicate Trididemnum solidum. J Nat Prod. 1988;51:1–21.
Gallo GG, Coronelli C, Vigevani A, Lancini GC. The structure of tetrenolin: a new antibiotic substance. Tetrahedron. 1969;25:5677–80.
Davidson BS, Ireland CM. Lissoclinolide, the first non-nitrogenous metabolite from a Lissoclinum tunicate. J Nat Prod. 1990;53:1036–8.
Richardson AD, Ireland CM. A profile of the in vitro antitumor activity of lissoclinolide. Toxicol Appl Pharmacol. 2004;195:55–61.
Miyairi N, Sakai HI, Konomi T, Imanaka H. Enterocin, a new antibiotic. J Antibiot. 1976;29:227–35.
Sitachitta N, Gadepalli M, Davidson BS. A new α-pyrone-containing metabolites from a marine-derived actinomycete. Tetrahedron. 1996;52:8073–80.
Kang H, Jensen PR, Fenical W. Isolation of microbial antibiotics from a marine ascidian of the genus didemnum. J Org Chem. 1996;61:1543–6.
Omura S, Iwai Y, Hirano A, Nakagawa A, Awaya J, Takahashi Y, et al. A new alkaloid AM-2282 of streptomyces origin. taxonomy, fermemtation, isolation and preliminary characterization. J Antibiot 1977;30:275–82.
Kinnel RB, Scheuer PJ. 11-hydroxystaurosporine: a highly cytotoxic, powerful protein kinase c inhibitor from a tunicate. J Org Chem. 1992;57:6327–9.
Strobel G, Li JY, Sugawara F, Koshino H, Harper J, Hess WM. Oocydin A, a chlorinated macrocyclic lactone with potent anti-oomycete activity from Serratia marcescens. Microbiology. 1999;145:3557–64.
Takada N, Sato H, Suenaga K, Arimoto H, Yamada K, Ueda K, et al. Isolation and structures of haterumalides NA, NB, NC, ND, and NA, novel macrolides from an okinawan sponge Ircinia sp. Tetrahedron Lett. 1999;40:6309–12.
Ueda K, Hu Y. Haterumalide B: a new cytotoxic macrolide from an okinawan ascidian Lissoclinum sp. Tetrahedron Lett. 1999;40:6305–8.
Bose U, Hodson MP, Shaw PN, Fuerst JA, Hewavitharana AK. Two peptides, cycloaspeptide A and nazumamide A from a sponge associated marine actinobacterium Salinispora sp. Nat Prod Commun. 2014;9:545–6.
Nishimura H, Katagiri K, Sato K, Mayama M, Shimaoka N. Toyocamycin, a new anti-candida antibiotics. J Antibiot.1956;9:60–62.
Zabriskie TM, Ireland CM. The isolation and structure of modified bioactive nucleosides from Jaspis johnstoni. J Nat Prod. 1989;52:1353–6.
Rohmer M, Ourisson G. Structure des bacteriohopanetetrols d’Acetobacter xylinum. Tetrahedron Lett. 1976;17:3633–6.
Costantino V, Fattorusso E, Imperatore C, Mangoni A. A biosynthetically significant new bacteriohopanoid present in large amounts in the Caribbean sponge Plakortis simplex. Tetrahedron. 2001;57:4045–8.
Sakai R, Higa T, Jefford CW, Bernardinelli G. Manzamine A, a novel antitumor alkaloid from a sponge. J Am Chem Soc. 1986;108:6404–5.
Waters AL, Peraud O, Kasanah N, Sims JW, Kothalawala N, Anderson MA, et al. An analysis of the sponge Acanthostrongylophora igens’ microbiome yields an actinomycete that produces that natural product manzamine A. Front Mar Sci. 2014;1:54.
Adamczeski M, Quinoa E, Crews P. Novel sponge-derived amino acids. 5. Structures, stereochemsitry, and synthesis of several new heterocycles. J Am Chem Soc. 1989;111:647–54.
Johnson TA, Sohn J, Vaske YM, White KN, Cohen TL, Vervoort HC, et al. Myxobacteria vs. sponge-derived alkaloids: the bengamide family identified as potent immune modulating agents by scrutiny of LC-MS/ELSD libraries. Bioorg Med Chem. 2012;20:4348–55.
Bewley CA, Debitus C, Faulkner DJ, Microsclerodermins A, Antifuncal B. Cyclic peptides from lithistid sponge Microscleroderma sp. J Am Chem Soc. 1994;116:7631–6.
Schmidt EW, Faulkner DJ. Microsclerodermins C–E, antifungal cyclic peptides from the lithistid marine sponges Theonella sp. and Microscleroderma sp. Tetrahedron. 1998;54:3043–56.
Hoffmann T, Müller S, Nadmid S, Garcia R, Müller R. Microsclerodermins from terrestrial myxobacteria: an intriguing biosynthesis likely connected to the sponge symbiont. J Am Chem Soc. 2013;135:16904–11.
Nicacio KJ, Ióca LP, Fróes AM, Leomil L, Appolinario LR, Thompson CC, et al. Cultures of the marine bacterium Pseudovibrio denitrificans Ab134 produce bromotyrosine-derived alkaloids previously only isolated from marine sponge. J Nat Prod. 2017;80:235–40.
Florean C, Kim KR, Schnekenburger M, Kim HJ, Moriou C, Debitus C, et al. Cell death induction by marine cytotoxin (+)-1(R), 6(S), 1’(R), 6’(S), 11(R), 17(S)-fistularin and Bcl-1 inhibitor venetoclax. Mar Drugs. 2018;16:E518.
Kernan MR, Cambie RC, Bergquist PR. Chemistry of sponges, VII. 11,19-dideoxyfistularin 3 and 11-hydroxyaerothionin, bromotyrosine derivatives from Pseudoceratina durissima. J Nat Prod. 1990;53:615–22.
Rodriguez AD, Piña IC. The structures of aplsinamisines I, II, and III; new bromotyrosine-derived alkaloids from the caribbean sponge Aplysina cauliformis. J Nat Prod. 1993;56:907–14.
Kobayashi J, Honma K, Sasaki T, Tsuda M, Purealidins J-R. New bromotyrosine alkaloids form the okinawan marine sponge Psammaplysilla purea. J Antibiot. 1995;43:403–7.
Kijjoa A, Bessa J, Wattanadilok R, Sawangwong P, Nascimento MSJ, Pedro M, et al. Dibromotyrosine derivatives, a maleimide, aplysamine-2 and other constituents of the marine sponge pseudoceratina purpurea. Z Naturforsch B Chem Sci. 2005;60:904.
Smith TE, Pond CD, Pierce E, Harmer ZP, Kwan J, Zachariah MM, et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat Chem Biol. 2018;14:179–85.
Ueoka R, Meodad RA, Gran-Scheuch A, Bhushan A, Fraaije MW, Piel J. Genome mining of oxidation modules in trans-acyltransferase polyketide synthases reveals a culturable soource of lobatamides. Angew Chem Int Ed. 2020;59:7761–5.
Crews P, Manes LV, Boehler M. Jasplakinolide, a cyclodepsipeptide from the marine sponge Jaspis sp. Tetrahedron Lett. 1986;27:2797–800.
Watts KR, Morinaka BI, Amagata T, Robinson SJ, Tenney K, Bray WM, et al. Biostructural features of additional jasplakinolide (Jaspamide) analogues. J Nat Prod. 2011;74:341–51.
Kunze B, Jansen R, Sasse F, Höfle G, Reichenbach H, Chondramides A-D. New antifungal and cytostatic depsipeptides from Chondromyces crocatus (Myxobacteria). J Antibiot. 1995;48:1262–6.
Iizuka T, Fudou R, Jojima Y, Ogawa S, Yamanaka S, Inukai Y, et al. Miuraenamides A and B, novel antimicrobial cyclic depsipeptides from a new slightly halophilic myxobacterium: taxonomy, production, and biological properties. J Antibiot. 2006;59:385–91.
Erickson KL, Beutler JA, Cardellina JH, Boyd MR. Salicylihalamides A and B, novel cytotoxic macrolides from the marine Sponge Haliclona sp. J Org Chem. 2001;66:1532.
Kunze B, Jansen R, Sasse F, Höfle G, Reichenbach H. Apicularens A and B, new cytostatic macrolides from Chondromyces species (Myxobacteria) production, physico-chemical and biological properties. J Antibiot. 1998;51:1075–80.
Kashman Y, Shmueli GU. Latrunculin, A new 2-thiazolidinone macrolide from the marine sponge Latrunculia magnifica. Tetrahedron Lett. 1980;21:3629–32.
Gerth K, Bedorf N, Höfle G, Irschik H, Reichenbach H. Epothilons A and B: antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria). Production, physico-chemical and biological properties. J Antibiot. 1996;49:560–3.
Acknowledgements
This work was supported in part by the National Institute of Health grant R01 1AI141511.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
McCauley, E.P., Piña, I.C., Thompson, A.D. et al. Highlights of marine natural products having parallel scaffolds found from marine-derived bacteria, sponges, and tunicates. J Antibiot 73, 504–525 (2020). https://doi.org/10.1038/s41429-020-0330-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-020-0330-5
This article is cited by
-
Challenges and Opportunities for Bioactive Compound and Antibiotic Discovery in Deep Space
Journal of the Indian Institute of Science (2023)
-
Comparative metabolomic analysis reveals shared and unique chemical interactions in sponge holobionts
Microbiome (2022)
-
Ancient plant-like terpene biosynthesis in corals
Nature Chemical Biology (2022)
-
Strategies to access biosynthetic novelty in bacterial genomes for drug discovery
Nature Reviews Drug Discovery (2022)
-
Marine fungal metabolites as a source of drug leads against aquatic pathogens
Applied Microbiology and Biotechnology (2022)