Abstract
The microbiome, of which the bacterial component alone (microbiota), is estimated to include 10 times more cells than human cells of the body, blooms immediately after birth and evolves in composition and complexity throughout childhood. The gut microbiome has a profound impact on gastrointestinal tract development, maintenance of mucosal surface integrity, and contributes to the nutritional status of the host and thus plays a pivotal role in health and disease. New technologies have enabled the detailed characterization of normal microbial symbionts and dysbiosis–disease associations. This review summarizes the stepwise establishment of the intestinal microbiota, influential environmental factors, and how this may be perturbed in preterm very-low-birth-weight infants. The contribution of the microbiota to provision of energy and nutrients for intestinal development and the nutritional status of the host are reviewed. In addition, the crucial role of the gut microbiota in maintaining mucosal integrity is explored along with how its breakdown can lead to sepsis, necrotizing enterocolitis, and systemic inflammatory response syndrome. Finally, the role of enteral feeding type (human milk, formula, and nutrient fortification) in mediating these processes is discussed, and guidance is provided for nutritional strategies to promote health in these fragile infants.
Similar content being viewed by others
Main
The microbiome describes the totality of the microbes in an environment, including bacteria, protozoa, viruses, fungi, and their genetic elements. The human microbiota describes the bacteria colonizing every surface of the body from the skin to the respiratory tract, genitourinary tract, and gastrointestinal tract (GIT). The intestinal microbiota constitutes the most abundant microbial community in humans with 10 times more cells and 150 times more genes than human cells. Until recently, little was known of the GIT microbiota of very-low-birth-weight (VLBW, <1,500 g) infants as traditional culture-based techniques identified only 20% of GIT microbes (1). Advances in molecular technologies have allowed a rapid expansion of knowledge of the human microbiome.
The GIT microbiota is crucial in maintaining mucosal integrity, and its breakdown can lead to sepsis, necrotizing enterocolitis (NEC), and systemic inflammatory response syndrome (2). Distal organ damage in systemic inflammatory response syndrome has been linked to severe morbidities associated with preterm birth including brain damage (periventricular leukomalacia) and lung damage (chronic lung disease) (2). The GIT microbiota best associated with optimal health is unknown; however, an overarching finding in VLBW infants is decreased GIT microbiota diversity and a higher pathogen load (1,3,4,5,6).
Select Approaches of Characterizing the Intestinal Microbiome
In the past century, investigations of the GIT microbiota were cultured based with the identification of the genus, species, and even strain using a battery of morphological and biochemical tests. However, up to 80% of gut microbes remain very difficult to culture (1). A number of new molecular-based approaches have been developed over the past decade revolutionizing the study of microbial communities. High-throughput sequencing of the 16S ribosomal RNA gene is the most widely employed approach used to characterize a microbial community. The 16S rRNA is part of the small ribosomal unit that is preserved in all organisms and contains highly conserved sequence domains interspersed with hypervariable regions. Selective amplification of these hypervariable regions followed by high-throughput sequencing is an efficient way to characterize the microbial community (7).
Beyond 16S rDNA–based sequencing, metagenomic, metatranscriptomic, metaproteomic, metabolomic, and non–16S rDNA-targeted approaches are increasingly employed to generate functional and quantitative information on microbial communities (8). The metagenome refers to the totality of the genomes from the whole community including bacteria, fungi, viruses, and protozoa. To analyze a metagenome, DNA is extracted and shotgun sequenced to determine functional and biochemical capabilities of the microbial community. Two strains of any given bacterial species may differ in DNA content by as much as 25% allowing for the same species but different strains to be harmless, commensal, or harmful.
A further step is to identify not only which genes are present in a microbiome but which genes have been expressed into mRNA (metatranscriptomics) or proteins (metaproteomics) (8). At present, metatranscriptomics of the GIT microbiome is limited by the inherent instability of RNA and difficulties extracting it from stool. Metaproteomics is an attractive alternative but has been limited because of analytical challenges, incomplete reference databases, and highly redundant protein sequences between different microbes. Metabolomic analyses use mass spectrometry or nuclear magnetic resonance spectrometry to monitor the whole set of small molecules produced by microbes or the host cells within a sample (8).
Normal Intestinal Colonization
Predominant gut organisms from the kingdom bacteria are presented in Figure 1 . More than 1,000 bacterial species have been identified in the adult GIT with the three phyla being prominent: Bacteroides, Actinobacteria, and Firmicutes. In any single individual, 99% of the GIT microbiota is made up of 30–40 species (9). Until recently, we believed that the GIT of a healthy-term newborn was sterile; however, meconium is now known to contain microbial rDNA (10), suggesting that infants are colonized before birth. Oral administration of genetically labeled Enterococcus faecium to pregnant mice led to their presence in the meconium of born pups by cesarean, demonstrating the possibility of prenatal maternal microbial transmission (11).
Classification of bacteria documented to colonize the gastrointestinal tract of very-low-birth-weight infants.
Immediately after birth, bacterial colonization of the GIT proceeds rapidly (phase I, 1–2 wk) with aerotolerant microbes. It continues through exclusive breastfeeding (phase II) until weaning (phase III) in a stepwise manner with the appearance of strict anaerobes such that by 18–24 mo of age, the microbiota resembles the extremely dense and complex microbial colonization of an adult (phase IV) (7,12,13). The infant microbiota is more variable day-to-day in its composition and less stable over time compared with that of the adult (1,12,14,15). The relative stability of the adult microbiota has been challenged by a recent study demonstrating that a single week of dietary change is sufficient to alter the intestinal microbiota (16). Infant microbial colonization can be significantly affected by exogenous factors including the mode of delivery, feeding type, antibiotics, and introduction of solid food.
Intestinal Colonization of the Preterm VLBW Infant
Colonization of the GIT is perturbed by a number of factors prevalent among VLBW infants including: cesarean delivery, antibiotics (mother or infant), prolonged rupture of the membranes, parenteral feeding, delayed enteral feeding, slower GIT transit time, gestational age, birth weight, living in a populated neonatal intensive care unit with an enriched pathogen load, and lack of exposure to mother’s skin and breast milk microbiome (1,3,4,5,6,13,17,18,19,20,21,22).
Table 1 summarizes studies describing the GIT microbiota of VLBW infants using molecular techniques. There appear to be significant differences in the composition of the intestinal microbiota of preterm compared to term infants, with decreased bacterial diversity, increased pathogens potentially related to NEC, and a surprising increase in eukaryotic and viral diversity (3,6,23,24,25,26,27,28,29,30). As described in both Tables 1 and 2 , these studies represent small numbers of patients and samples with between 1 and 29 patients per study in the VLBW group and between 3 and 18 patients in the NEC group. Other limiting factors in studying these data are the heterogeneity (or lack of reporting) of patient characteristics, diet, mode of delivery, antibiotic course, and sampling frequency and timing (some samples only collected after the onset of NEC).
Impact of Mode of Delivery
Infants born by cesarean have a different GIT microbiome compared with those born vaginally, and some of these differences are sustained throughout early childhood (3,14,15,20,31,32). Vaginal birth leads to an inoculation of infants with maternal fecal and vaginal bacteria. In contrast, cesarean-born infants are exposed initially to bacteria originating from the hospital environment, health care workers, and their mother’s skin. In term infants, vaginal delivery results in a higher GIT bacterial richness (number of different species) and diversity (richness and evenness of their distribution) (20,31). While study-to-study differences exist, most recently, Azad et al. (31) reported that Escherichia-Shigella is underrepresented, and the phylum Bacteroidetes was undetectable in 4-mo-old term-born infants born by cesarean compared with vaginal delivery. Contrary to previous studies, this group did not observe differences by mode of delivery in prevalence of Clostridium difficile or the relative abundance of Bifidobacterium or Clostridium.
Impact of Antibiotic Exposure
Most VLBW infants receive broad-spectrum antibiotics during their early postnatal course possibly resulting in inadequate phase I colonization (13) with an apparent inverse correlation between the number of days of antibiotics in the first month postnatally and microbial diversity as well as total bacterial load in stools (18). Early studies suggested that colonization with beneficial bacteria such as Lactobacillus is specifically affected (19,33) and that antibiotic usage promotes a bloom of Staphylococcus. More recently, it was shown term infants treated with parenteral ampicillin and gentamicin within the first 48 h of birth demonstrated significant reductions in the phyla Actinobacteria (including Bifidobacterium) and Firmicutes (including Lactobacillus) which were replaced by Proteobacteria (including Enterobacteriaceae) (34). The dominance of Proteobacteria and reduced microbial diversity remained for at least 8 wk after treatment. In an adult human study, exposure to clindamycin for 7 d led to a decline in bacterial diversity that persisted for up to 2 y (35). Given the instability of the newborn GIT microbiota, antibiotics exposure could be expected to induce profound alterations of the microbial community with long-term consequences (27,29,35).
Impact of Diet
Healthy-term newborns appear to be initially colonized by large numbers of Enterobacter and Streptococcus, regardless of feeding type (12). It is proposed that these bacteria are responsible for creating a reduced environment in the GIT which favors establishment of the anaerobes Bacteroides, Bifidobacterium, and Clostridium by day 4 to 7. In the term breastfed infant, it has been shown that by 7 d, the beneficial Bifidobacteria and Lactobacillus predominate (36). In formula-fed infants, a more adult-type flora is most prevalent (13,32,37,38) along with an abundance of potentially pathogenic bacteria (Clostridium difficile and Escherichia coli) (5,36,39). Most recently, Azad et al. (31) reported lower bacterial richness and diversity in the stools of 4-mo-old breastfed infants compared with formula-fed infants; further formula-fed infants had increased richness and overrepresentation of C. difficile compared with breastfed infants.
The differences in colonization of human milk-fed compared with formula-fed infants are believed due, in part, to breast milk inoculating the GIT with its own rich microbiome and abundant source of oligosaccharides that selectively stimulate the growth and/or activity of beneficial bacteria (40,41). It has been shown that during breastfeeding a significant volume of milk flows from the baby’s mouth back into the mammary ducts providing an opportunity for bacteria from the baby’s mouth to make its way into breast milk (42). Even after cleansing the breast using an iodine solution to minimize bacterial contamination, pyrosequencing of the 16S rRNA gene from milk reveals the microbial community in human milk to be highly diverse and complex (40,43). Data from human and animal studies suggest that bacteria or their components may actively migrate from the maternal GIT, by way of macrophages or dendritic cells, to the mammary gland and breast milk (21,44), thereby providing another mechanism for different colonization patterns of human milk-fed compared with formula-fed infants. Preliminary data also suggest that maternal obesity and elective cesarean section may also be associated with microbial diversity and composition in breast milk (40).
Role of the Microbiome on GIT Development and Maintenance of Mucosal Surface Integrity
Human beings are born with a naive immune system that must develop tolerance to its environment and mature to develop an appropriate immunological response to pathogens. The GIT represents the largest surface area exposed to the external environment and contains 70–80% of the body’s immune cells (45). The microbiota communicates with host cells via Toll-like receptors (TLR) which are transmembrane proteins present throughout the GIT that sense microbes by their conserved molecular patterns. For example, TLR-2 recognizes peptidoglycan and lipoteichoic acid from Gram-positive bacteria, TLR-4 recognizes lipopolysaccharide, the endotoxin of Gram-negative bacteria, and TLR-5 recognizes flagellin (46). Paradoxically, signaling induced by commensal bacteria is required for development of gut epithelial cell protection against inflammatory mediator injury as well as for cell repair (47). The effects of commensal bacteria include enhancement of tight junctions, stimulation of mucin production, and downregulation of cytokine production. Evidence exists for induction of enterocyte protection by Lactobacillus, Bifidobacterium, Bacteroides thetaiotaomicron, and Streptococcus thermophiles (46).
The microbiota may impact not only epithelial cell development but also endothelial cell ontogenesis. Intestinal angiogenesis is ongoing at the time of acquisition of the microbiome, and a perturbation of angiogenesis may be implicated in intestinal inflammatory conditions such as NEC. In a laboratory model, fibroblasts have been shown to produce proangiogenic factors in response to microbial cell products, and endothelial cells have demonstrated proliferation, migration, tube formation, and vessel sprouting (48).
Further evidence for the role of the microbiome on gastrointestinal development comes from experiments with germ-free animals as well as from alteration of the microbiome with antibiotics. Germ-free rodents have a decrease in transcription of mucin genes and a decreased thickness of the mucus layer along with fewer Goblet cells and less IgA production (22). Suckling rats exposed to 2 wk of antibiotics at the time of weaning show significant downregulation of genes encoding Paneth cell products (important in host defense) and the major histocompatibility complex Ib and II proteins (antigen presentation), thus potentially altering the development of tolerance to food antigens (22).
The Energy and Nutritient Contribution of the GIT Microbiota
Several lines of evidence suggest that the microbiota of the distal small intestine and colon make a substantial contribution to the nutrition of the host (up to 40–50% of energy intake). Germ-free animals have a higher dietary requirement for energy than those with an intact microbiota and when the microbiota of conventionally reared mice are transplanted into to lean germ-free mice, their body weight increases dramatically (49). Many bacterial species in the distal intestine can ferment undigested carbohydrates (e.g., breast milk oligosaccharides) to produce the short-chain fatty acids acetate, propionate, and butyrate (50). Butyrate is a major energy source for colonocytes, and propionate and acetate are absorbed and enter the portal circulation where they are used by a variety of body tissues, particularly the liver, in energy metabolism, lipogenesis, and gluconeogenesis. Additionally, short-chain fatty acids are believed to influence energy metabolism by binding to and activating G protein-coupled receptors on colonic epithelial and enteroendocrine cells resulting in the release of peptide YY and glucagon-like peptide. We reported piglets fed formulas containing a mixture of inulin (fructooligosaccharide source) and galactooligosaccharides for the first 28 d had colons that weighed ~40% more than piglets fed the same formulas but without a source of oligosaccharides (51). Increased bacterial load supported by the provision of undigested carbohydrates and the resultant microbial synthesis of butyrate is thought to play a role in increased intestinal cell proliferation. These findings highlight the importance of the microbiota for the VLBW infant with increased energy requirements whose colon must double in length before reaching 40 wk corrected age.
Differences in microbial community composition between normal weight and obese children and adults have been observed (50). Important in reference to the VLBW infant, new data from developing countries suggest that the GIT microbial composition is altered in both marasmus (malnutrition due to insufficient energy) and kwashiorkor (malnutrition due to insufficient protein) (50). Interestingly, transplantation of the microbiota of Malawian children with kwashiorkor to gnotobiotic mice fed a rodent diet patterned after the Malawian diet resulted in significant perturbation of both amino acid and carbohydrate metabolism (52).
It has long been recognized that intestinal bacteria are critical in synthesizing a number of vitamins, vitamin K being the classic example. Routine vitamin K prophylaxis of healthy newborns is required to prevent hemorrhagic disease as a result of minimal placental transfer of the vitamin but also due to the low concentrations of vitamin K producing bacteria in the GIT at birth. Our own work in this area suggests that the total amount of the B-vitamin folate synthesized by bacteria in the colon may approach or even exceed dietary intake in both infants and adults and it can be absorbed (53,54). Folate plays an important role in DNA and RNA biosynthesis, amino acid synthesis, and cell division, all processes important for healthy GIT development. Other vitamins thought to be synthesized by bacteria include vitamin B12, biotin, thiamine, riboflavin, and pyridoxine.
Although beyond the scope of this review, the GIT microbiota also contributes a wide variety of other small molecules which have important functions. These metabolites include bile acids (facilitate fat absorption), choline metabolites (modulate lipid and glucose homeostasis), phenolic, benzoyl, and phenyl derivatives (detoxification of xenobiotics), indole derivatives (protect against stress-induced lesions in the gut), lipids, and many more (55).
Impact of the Microbiome on Health Outcomes in VLBW Infants
Short Term
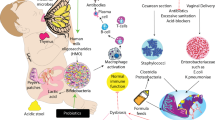
The interaction between the VLBW infant and their microbiome is presented in Figure 2 . The VLBW baby is at elevated risk for feeding intolerance, NEC, and sepsis. The pathophysiology of these disorders is likely multifactorial involving a combination of intestinal mucosal barrier immaturity, an imbalance in microvascular tone, aberrant microbial colonization and an unbalanced immune response (56). Infectious causes for NEC have been sought for more than 30 years with the implication of several bacteria (namely several species of Clostridium, Klebsiella pneumoniae, Escherichia coli, and Cronobacter sakazakii) and viruses (namely Coronavirus, Coxsackie, Rotavirus, Adenovirus, and Torovirus) (8). With the advent of molecular technologies, it has become possible to not only study the bacterial species present but also the metabolic differences in the GIT microbiota of infants with and without NEC.
Impact of preterm birth on the gastrointestinal microbiota. IBD, inflammatory bowel disease; NEC, necrotizing enterocolitis; SIRS, systemic inflammatory response syndrome.
Table 2 summarizes the molecular studies of GIT microbiota for VLBW infants with NEC or sepsis. In summary, these studies demonstrate lower bacterial diversity, less Bifidobacteria and Bacteroidetes, and a bloom of the γ-Proteobacteria 1–3 wk prior to the diagnosis of NEC (57,58,59,60,61,62). A metagenomic analysis on stool samples from a set of twins discordant for the diagnosis of NEC demonstrated significant differences in the genes mapping to carbohydrate metabolism leading to speculation that microbial communities may metabolize milk through different pathways resulting in differing substrates available to the microbiota and thus differential effects on the host (57). Similar perturbations in the GIT microbiota development have been reported for infants with late-onset sepsis (29).
The GIT microbiome plays a pivotal role in balancing the inflammatory system in the immature GIT. A breakdown of the mucosal barrier can result in translocation of inflammatory mediators producing systemic inflammatory response syndrome. In addition to sepsis and NEC, systemic inflammatory response syndrome plays a key role in downstream organ inflammation and damage that may result in severe long-term morbidities such as periventricular leukomalacia, chronic lung disease, and retinopathy of prematurity (2).
Long Term
The VLBW infant is at elevated risk of metabolic syndrome including type II diabetes, hypertension, and obesity in later life. This may commence with an altered growth during the neonatal period related to suboptimal nutritional intake, illness, and potentially an aberrant microbiota with altered metabolic capacity, resulting in a greater percentage of body fat and considerably less lean mass at term-corrected age (63,64). Evidence for the role of the microbiota in obesity comes from rodent studies with induction of hepatic lipogenesis and increased lipid storage in adipocytes of germ-free mice after colonization with normal gut microbiota (65). In human twin studies, evidence suggests that physiological phenotype such as adiposity may be better associated with a core microbiome at the gene level rather than at the organismal lineage level with a decrease in bacterial diversity and an alteration in metabolically active genes found in obese patients (66).
Dysregulation of the developing immune system has been implicated in a wide array of diseases in childhood and adult life including atopy (67) and food allergy (68), inflammatory bowel disease (69), and cancer (70). Animal studies have demonstrated that both changes in diet and in the gut microbiota alter the development of autoantibodies to pancreatic β-cells, a precursor to the development of type I diabetes (71), but whether these translate to human conditions remains speculative.
An emerging area of interest is the impact of the gut microbiota on the developing human brain which may be of particular importance for the preterm infant who is at elevated risk for behavioral issues including attention-deficit disorder and autism spectrum disorder. The central nervous system communicates bidirectionally with the gut via the enteric nervous system. Studies in germ-free mice have revealed important changes in neurotrophin factors related to brain plasticity as well as in behavior with increased stress responses and anxiety-like behavior. These abnormal behaviors can be reversed with transplantation of a normal gut microbiota (45).
Feeding Recommendations to Support a Healthy GIT Microbiome
The portfolio of evidence indicates that mother’s own milk is the optimal way to feed every infant, including VLBW infants. This principle is internationally endorsed based on extensive literature, primarily derived from studies with healthy-term infants, associating provision of own mother’s milk with decreased incidence of diarrhea, otitis media, hospitalization for lower respiratory tract infection in the first week of life, sudden infant death syndrome, improved neurodevelopment, and perhaps decreased risk of type II diabetes and a small decrease in childhood obesity (72,73,74). Additionally, among VLBW infants, provision of own mother’s milk is associated with lower incidence of NEC and sepsis, improved feeding tolerance, and a reduction in colonization by pathogens.
For VLBW infants at risk of NEC, when own mother’s milk is not available, pasteurized donor human milk is recommended. A Cochrane review reported a higher incidence of NEC (relative risk of 2.5 (95% confidence interval: 1.2, 5.1)) and feeding intolerance (relative risk: 4.9 (1.7, 20.7)) among infants supplemented with formula compared with donor human milk (75). In North America, donor milk provided to VLBW infants is pasteurized to remove the possibility of transmission of harmful agents including HIV and known pathogens. Pasteurization, however, destroys all live cells, and hence inoculation of the infant gut with bacteria may be diminished. However, pasteurized donor milk remains a rich source of oligosaccharides which are seemingly unaffected by heat treatment (76).
In order to meet the nutritional requirements of VLBW infants, it is standard of care to add energy and essential nutrients to human milk after tolerance to enteral feeds has been established. There is strong evidence that early nutrient deficits and suboptimal growth, common in the neonatal intensive care unit, are independent risk factors for poor VLBW outcome (63). Human milk for the majority of VLBW infants is fortified using nutrient fortifiers whose protein source is bovine based although a human milk–derived fortifier is commercially available. The impact of human milk fortification on the gut microbiota has not been systematically investigated, although two industry-sponsored trials have demonstrated at least a 50% reduction in NEC and NEC requiring surgical intervention was almost eliminated (77,78). Unfortunately, the study design does not allow for determination of whether it was the absence of bovine-based fortifier, formula, or both that reduced the incidence of NEC.
A variety of probiotics have been studied in preterm infants, most commonly Lactobacillus and Bifidobacterium. A Cochrane meta-analysis of 20 randomized or quazi-randomized trials (n = 5,529 infants) reported that probiotic supplementation of preterm infants reduced both all-cause mortality (relative risk: 0.65 (0.52, 0.81)) and NEC (relative risk: 0.43 (0.33, 0.56)) (79). Furthermore, probiotic supplementation improved the antecedent of severe NEC, feeding intolerance, as well as reduced the number of days of hospitalization. Unlike in Europe and other parts of the world, probiotic supplementation has not been widely adopted as a NEC prevention strategy in North American neonatal intensive care units due to concerns about the limited safety data for infants born <1,000 g and a domestically available probiotic formulation with demonstrated efficacy, safety, and regulatory approval. There remains some concerns that the regulatory framework for approval of probiotics does not approach the rigor for drugs. The Canadian Pediatric Society Committee recently recommended that physicians consider recommending probiotics for the prevention of NEC for at-risk preterm infants (80). An in-depth understanding of the microbial community composition and metabolic functions in the GIT of the “well” VLBW infant fed mother’s own milk will leverage the incorporation into clinical practice of therapeutics such as probiotics because at a very practical level, we will know what the “gold standard” is for microbial composition and function (55).
Conclusions
A growing body of evidence suggests that perturbed colonization of the microbiota in the preterm VLBW infant is associated with increased morbidity and mortality during initial hospitalization and beyond. The potential to develop strategies to reduce morbidities associated with very preterm birth through manipulation of the microbiome appear tremendous. While the microbial community composition and related metabolic functions best associated with optimal health in VLBW infants are unknown, characterization of the GIT microbiome of the “well” exclusively own mother’s milk-fed VLBW infant seems a reasonable starting point. High-throughput sequencing of the 16S ribosomal RNA gene has already begun to yield critical information on the microbial community in the GIT of the VLBW infant, and newer emerging technologies including metatranscriptomics, metaproteomics, and metabolomics show much promise in the future of helping to define the complex metabolic environment in the GIT. This will facilitate a sophisticated understanding of the microbiome in relation to optimal health and provide a basis against which to assess novel nutritional therapies.
Statement of Financial Support
This study was supported through a grant from the Canadian Institutes for Health Research, MRC Operating Grant Program (MOP) 210093 and 125997.
Disclosures
No disclosures.
References
Mshvildadze M, Neu J . The infant intestinal microbiome: friend or foe? Early Hum Dev 2010;86:Suppl 1:67–71.
Neu J, Douglas-Escobar M, Lopez M . Microbes and the developing gastrointestinal tract. Nutr Clin Pract 2007;22:174–82.
Brooks B, Firek BA, Miller CS, et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2014;2:1.
Cotten CM, Taylor S, Stoll B, et al.; NICHD Neonatal Research Network. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics 2009;123:58–66.
Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006;118:511–21.
Schwiertz A, Gruhl B, Löbnitz M, Michel P, Radke M, Blaut M . Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res 2003;54:393–9.
Mshvildadze M, Neu J, Mai V . Intestinal microbiota development in the premature neonate: establishment of a lasting commensal relationship? Nutr Rev 2008;66:658–63.
Morowitz MJ, Poroyko V, Caplan M, Alverdy J, Liu DC . Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics 2010;125:777–85.
Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635–8.
Moles L, Gómez M, Heilig H, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 2013;8:e66986.
Jiménez E, Marín ML, Martín R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol 2008;159:187–93.
Mackie RI, Sghir A, Gaskins HR . Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 1999;69:1035S–45S.
Sherman PM, Cabana M, Gibson GR, et al. Potential roles and clinical utility of prebiotics in newborns, infants, and children: proceedings from a global prebiotic summit meeting, New York City, June 27-28, 2008. J Pediatr 2009;155:S61–70.
Grönlund MM, Lehtonen OP, Eerola E, Kero P . Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr 1999;28:19–25.
Salminen S, Gibson GR, McCartney AL, Isolauri E . Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 2004;53:1388–9.
David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63.
Fanaro S, Chierici R, Guerrini P, Vigi V . Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 2003;91:48–55.
Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P . Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed 1999;80:F167–73.
Hall MA, Cole CB, Smith SL, Fuller R, Rolles CJ . Factors influencing the presence of faecal lactobacilli in early infancy. Arch Dis Child 1990;65:185–8.
Huurre A, Kalliomäki M, Rautava S, Rinne M, Salminen S, Isolauri E . Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology 2008;93:236–40.
Perez PF, Doré J, Leclerc M, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 2007;119:e724–32.
Schumann A, Nutten S, Donnicola D, et al. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genomics 2005;23:235–45.
Arboleya S, Ang L, Margolles A, et al. Deep 16S rRNA metagenomics and quantitative PCR analyses of the premature infant fecal microbiota. Anaerobe 2012;18:378–80.
Arboleya S, Binetti A, Salazar N, et al. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 2012;79:763–72.
Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA . Microbiome assembly across multiple body sites in low-birthweight infants. MBio 2013;4:e00782–13.
LaTuga MS, Ellis JC, Cotton CM, et al. Beyond bacteria: a study of the enteric microbial consortium in extremely low birth weight infants. PLoS One 2011;6:e27858.
Madan JC, Salari RC, Saxena D, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed 2012;97:F456–62.
Magne F, Abély M, Boyer F, Morville P, Pochart P, Suau A . Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol 2006;57:128–38.
Mai V, Torrazza RM, Ukhanova M, et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One 2013;8:e52876.
Morowitz MJ, Denef VJ, Costello EK, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA 2011;108:1128–33.
Azad MB, Konya T, Maughan H, et al.; CHILD Study Investigators. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ 2013;185:385–94.
Sakata H, Yoshioka H, Fujita K . Development of the intestinal flora in very low birth weight infants compared to normal full-term newborns. Eur J Pediatr 1985;144:186–90.
Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM . The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin Nutr 2006;25:361–8.
Fouhy F, Guinane CM, Hussey S, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 2012;56:5811–20.
Jernberg C, Löfmark S, Edlund C, Jansson JK . Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007;1:56–66.
Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 2000;30:61–7.
Balmer SE, Wharton BA . Diet and faecal flora in the newborn: breast milk and infant formula. Arch Dis Child 1989;64:1672–7.
Stark PL, Lee A . The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol 1982;15:189–203.
Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE . Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett 2005;243:141–7.
Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A . The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 2012;96:544–51.
Newburg DS, Neubauer SH . Carbohydrates in milks: analysis, quantities and significance. In: Jensen RG, ed. Handbook on Milk Composition. San Diego, CA: Academic Press, 1995:273–349.
Ramsay DT, Kent JC, Owens RA, Hartmann PE . Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 2004;113:361–7.
Hunt KM, Foster JA, Forney LJ, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One 2011;6:e21313.
Jeurink PV, van Bergenhenegouwen J, Jiménez E, et al. Human milk: a source of more life than we imagine. Benef Microbes 2013;4:17–30.
Douglas-Escobar M, Elliott E, Neu J . Effect of intestinal microbial ecology on the developing brain. JAMA Pediatr 2013;167:374–9.
Torrazza RM, Neu J . The altered gut microbiome and necrotizing enterocolitis. Clin Perinatol 2013;40:93–108.
Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R . Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004;118:229–41.
Schirbel A, Kessler S, Rieder F, et al. Pro-angiogenic activity of TLRs and NLRs: a novel link between gut microbiota and intestinal angiogenesis. Gastroenterology 2013;144:613–623.e9.
Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004;101:15718–23.
Ramakrishna BS . Role of the gut microbiota in human nutrition and metabolism. J Gastroenterol Hepatol 2013;28:Suppl 4:9–17.
Aufreiter S, Kim JH, O’Connor DL . Dietary oligosaccharides increase colonic weight and the amount but not concentration of bacterially synthesized folate in the colon of piglets. J Nutr 2011;141:366–72.
Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013;339:548–54.
Aufreiter S, Gregory JF 3rd, Pfeiffer CM, et al. Folate is absorbed across the colon of adults: evidence from cecal infusion of (13)C-labeled [6S]-5-formyltetrahydrofolic acid. Am J Clin Nutr 2009;90:116–23.
Kim TH, Yang J, Darling PB, O’Connor DL . A large pool of available folate exists in the large intestine of human infants and piglets. J Nutr 2004;134:1389–94.
Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science 2012;336:1262–7.
Neu J, Walker WA . Necrotizing enterocolitis. N Engl J Med 2011;364:255–64.
Claud EC, Keegan KP, Brulc JM, et al. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 2013;1:20.
de la Cochetiere MF, Piloquet H, des Robert C, Darmaun D, Galmiche JP, Roze JC . Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: the putative role of Clostridium. Pediatr Res 2004;56:366–70.
Millar MR, Linton CJ, Cade A, Glancy D, Hall M, Jalal H . Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J Clin Microbiol 1996;34:2506–10.
Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N, Mai V . Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr 2010;156:20–5.
Torrazza RM, Ukhanova M, Wang X, et al. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PLoS One 2013;8:e83304.
Wang Y, Hoenig JD, Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J 2009;3:944–54.
Lucas A . Long-term programming effects of early nutrition – implications for the preterm infant. J Perinatol 2005;25:Suppl 2:S2–6.
Plagemann A, Harder T, Schellong K, Schulz S, Stupin JH . Early postnatal life as a critical time window for determination of long-term metabolic health. Best Pract Res Clin Endocrinol Metab 2012;26:641–53.
Reinhardt C, Reigstad CS, Bäckhed F . Intestinal microbiota during infancy and its implications for obesity. J Pediatr Gastroenterol Nutr 2009;48:249–56.
Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4.
Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 2007;56:661–7.
Kim JS, Sampson HA . Food allergy: a glimpse into the inner workings of gut immunology. Curr Opin Gastroenterol 2012;28:99–103.
Young VB . The intestinal microbiota in health and disease. Curr Opin Gastroenterol 2012;28:63–9.
Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 2013;105:1907–11.
Vaarala O . Is the origin of type 1 diabetes in the gut? Immunol Cell Biol 2012;90:271–6.
World Health Organization. Global strategies for infant and young child feeding, 2003. (http://www.who.int/nutrition/publications/gs_infant_feeding_text_eng.pdf).
Section on Breastfeeding. Breastfeeding and the use of human milk. Pediatrics 2012;129:e827–41.
Kim J, Unger S . Human milk banking. Paediatr Child Health 2010;15:595–602.
Quigley MA, Henderson G, Anthony MY, McGuire W . Formula milk versus donor breast milk for feeding preterm or low birth weight infants. Cochrane Database Syst Rev 2007:CD002971.
Bertino E, Coppa GV, Giuliani F, et al. Effects of Holder pasteurization on human milk oligosaccharides. Int J Immunopathol Pharmacol 2008;21:381–5.
Cristofalo EA, Schanler RJ, Blanco CL, et al. Randomized trial of exclusive human milk versus preterm formula diets in extremely premature infants. J Pediatr 2013;163:1592–1595.e1.
Sullivan S, Schanler RJ, Kim JH, et al. An exclusively human milk-based diet is associated with a lower rate of necrotizing enterocolitis than a diet of human milk and bovine milk-based products. J Pediatr 2010;156:562–7.e1.
AlFaleh K, Anabrees J . Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev 2014;4:CD005496.
Marchand V . Using probiotics in the paediatric population. Paediatr Child Health 2012;17:575–6.
Author information
Authors and Affiliations
Corresponding author
PowerPoint slides
Rights and permissions
About this article
Cite this article
Unger, S., Stintzi, A., Shah, P. et al. Gut microbiota of the very-low-birth-weight infant. Pediatr Res 77, 205–213 (2015). https://doi.org/10.1038/pr.2014.162
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2014.162
This article is cited by
-
Epidemiology, risk factors, clinical presentation and complications of late-onset neonatal sepsis among preterm neonates in Cyprus: a prospective case-control study
BMC Pediatrics (2024)
-
Composition and evolution of the gut microbiota of growing puppies is impacted by their birth weight
Scientific Reports (2023)
-
Early gut microbiota in very low and extremely low birth weight preterm infants with feeding intolerance: a prospective case-control study
Journal of Microbiology (2022)
-
Maturation of the preterm gastrointestinal tract can be defined by host and microbial markers for digestion and barrier defense
Scientific Reports (2021)
-
Intestinal tract and parenteral multi-organ sequential pathological injury caused by necrotizing enterocolitis
BMC Pediatrics (2020)