Abstract
Introduction:
Appeal for the domestic pig as a preclinical model for neurodevelopmental research is increasing. One limitation, however, is lack of magnetic resonance imaging (MRI) methods for brain volume quantification in the neonatal piglet. The purpose of this study was to develop and validate MRI methods for estimating brain volume in piglets.
Results:
The results showed that MRI and manual segmentation reliably estimated the changes in volume of different brain regions in 2- and 5-wk-old piglets. Substantial increases in the volumes of all brain regions examined were evident during the 3-wk period.
Discussion:
MRI can provide accurate estimates of brain region volume during the neonatal period in piglets. A piglet model that can be used in longitudinal studies may be useful for investigating how experimental (e.g., nutrition, infection) factors affect brain growth and development.
Methods:
Anatomic MRI data (non-longitudinal) were acquired 2- and 5-wk-old piglets using a three-dimensional T1-weighted magnetization-prepared gradient echo (MPRAGE) sequence on a MAGNETOM Trio 3T imager. Manual segmentation was performed for volume estimates of total brain, cortical, diencephalon, brainstem, cerebellar, and hippocampal regions. The MRI-based hippocampal volume estimates in 2- and 5-wk-old piglets were validated using histological techniques and the Cavalieri method.
Similar content being viewed by others
Main
The domestic pig (Sus scrofa), due to its anatomic and physiologic similarities to humans, is a common, well-accepted preclinical model in cardiovascular, metabolic, and pediatric nutrition research (1,2,3). Because pigs and humans share similar patterns in brain growth and development, appeal for the pig as a model for neurodevelopmental research has recently increased (4,5). In both pigs and humans, the major brain growth spurt extends from late prenatal to early postnatal, which is different from other common animal models (6). Both pigs and humans are born with a brain weight that is roughly 25% of the final adult weight (6). In addition, gross anatomical features, including gyral pattern and distribution of gray and white matter of the neonatal pig brain, are similar to those of human infants (7,8). Furthermore, because the pig is a precocial species with fairly well-developed sensory and motor systems at birth, it is amenable to behavioral testing to assess cognitive development as early as 1–2 wks of age (9). Thus, the neonatal piglet is a potential preclinical translational model for investigating the effects of early-life experiential factors (e.g., infection, nutrition) on brain and cognitive development. As the piglet is already used to study parenteral and enteral nutrition in preterm infants (10), and preterm birth is associated with a high rate of developmental delay and cognitive disability (11), the availability of the piglet for investigating brain and cognitive development would be particularly welcome. One limitation, however, has been the ability to make use of quantitative magnetic resonance imaging (MRI) for in vivo assessment of brain growth and development in neonatal piglets.
Quantitative MRI provides important information on brain development in early childhood and adolescence (12,13,14,15,16), but because of the potential health concerns for infants, few studies have focused on the period from birth to 4 y of age, when dramatic brain development occurs. Most MRI studies of infants have been with those born prematurely or with significant health complications (17,18,19). Recently, MRI has been used to assess the structural brain development of healthy full-term infants from birth to 2 y of age (13). In the first year after birth, total brain volume more than doubled, and hemispheric gray and white matter increased 149 and 11%, respectively. Although this application of MRI in human infants is impressive, the ability to address the mechanistic questions related to experiential influences on brain growth and development is not adequate at present because of practical or ethical concerns.
In older pigs, it is possible to obtain reliable estimates of brain volumes using MRI (20), and MRI has been used in experimental neurosurgical procedures and to investigate several neuropathological conditions (21,22,23,24,25). To the best of our knowledge, however, MRI has not been used to obtain brain volumetric data for in vivo quantitative analysis in neonatal piglets. As a first step toward addressing this void, here, we report MRI scanning and analysis protocols for estimating total brain, cortex, diencephalon, cerebellum, hippocampus, and brainstem volumes in neonatal piglets at 2 and 5 wks of age. The reliability of the MRI-based volume estimates was assessed for the hippocampus by the Cavalieri method and planimetric analysis with physical histological sections obtained postmortem. The results indicate that MRI can provide an accurate in vivo estimate of the changes in volume of brain regions over time during the neonatal period. Furthermore, substantial increases in volumes for all brain regions examined were evident during the 3-wk period. The potential to quantify brain growth and development in vivo using MRI in a longitudinal study design makes the piglet an attractive preclinical animal model.
Results
Total brain and subregion volumes of 2- and 5-wk-old piglets (n = 6) are shown in Table 1 . As expected, there was a main effect of age on total brain, cortex, diencephalon, cerebellum, hippocampus, and brainstem volumes, demonstrating considerable increases in volume between the 2-wk and 5-wk periods. There was no main effect of sex on any of the brain regions studied, indicating that the volumes were similar in males and females at this age (P > 0.05 for all measures). Because of this, data for males and females (n = 6) were combined. Volume of the left and right hippocampal formation was similar within each animal irrespective of sex or age. Because no asymmetries were observed, the left and right hippocampal volumes were combined to create a total hippocampal volume measure at each age. A three-dimensional (3D) segmented brain including all of the aforementioned brain regions can be seen in Figure 1 .
To determine the reliability of the MRI-based volume estimates, the volume of the hippocampus was also determined from physical sections obtained postmortem. The hippocampus was chosen because it was smallest of all the areas examined and most difficult to manually segment. Furthermore, technical challenges prevented the whole-brain slicing at 5-µm intervals. Using the histological sections, segmentation of individual hippocampal subregions (dentate, CA1, and CA3) was possible; however, owing to the small size of the hippocampus in the young pig, it was not possible to identify these hippocampal subregions by MRI. Thus, only whole hippocampal volume was compared between the two methods. To reduce variance due to genetic variability, piglets from the same litter were used for the histological validation. A total of four piglets were used: two that were 2 wks of age and two that were 5 wks of age. The average volumes determined histologically at 2 and 5 wks of age were 165.29 mm3 (±9.75) and 246.70 mm3 (±11.74), respectively. The average volumes determined using MRI at 2 and 5 wks of age were 487.83 mm3 (±38.28) and 658.87 mm3 (±47.25), respectively. Although tissue blocks were not measured before processing, the difference in the volumes determined histologically and using MRI represents a reduction in the total volume (i.e., shrinkage) of ~64%. Finally, a strong correlation between hippocampal volumes determined by MRI and histology was evident (r = 0.9878, P = 0.0122).
The median intra- and interobserver agreement for two independent raters for MRI-based volume estimates is shown in Table 2 . Intraobserver agreement was consistent across brain regions, with the median values ranging from 94.4 to 96.6%. The coefficients of variation ranged from 2.1 to 6.4% across brain regions. The interobserver agreement medians ranged from 96.6 to 98.8%, with coefficients of variation ranging from 2.0 to 5.2%.
Discussion
MRI has been used in older pigs to quantify whole-brain and substructure volumes; however, to the best of our knowledge, no studies have used MRI to characterize normal brain development during the neonatal period (20). Therefore, this study sought to determine whether MRI could be used to estimate the volumes of different brain regions in the neonatal period. The important results showed that MRI and manual segmentation reliably estimated the changes in the volume of different brain regions over time. Furthermore, substantial increases in the volumes of all the brain regions examined were evident during the 3-wk period.
Several challenges are presented when using MRI during the neonatal period, including poor spatial resolution and low tissue contrast, which cause difficulties in discerning between gray matter, white matter, and cerebrospinal fluid (26). Because of this, using automated segmentation procedures in the neonate has been difficult. An additional obstacle of structural analysis in the neonatal pig brain is the lack of a brain atlas specific to this age. Currently, the only MRI-based atlas for the pig is in adult animals, which is not suitable for neonatal brain analysis (27). Because there is currently no atlas available for pigs during the neonatal period, we chose to use manual segmentation for volume analysis. This technique requires significantly more time than semi- and fully automated segmentation methods, but provides accurate volume estimations. Additional imaging data sets are needed for the creation of structural MRI atlases, which will allow for more complex, automated segmentation methods. A future goal is to utilize more sophisticated techniques used for human studies, such as neonatal cortical mapping, once age-based MRI atlases have been created for piglets (17). Nonetheless, the current procedure can be used in a longitudinal experimental design to assess the effects of experiential factors (e.g., nutrition, infection) on brain growth and development. This is important because, similar to humans (6), overall brain weight of piglets in the neonatal period is about 25–35% of normal adults.
Studies employing MRI for the volumetric analysis of specific brain regions typically validated this modality using classic histological techniques and the Cavalieri method (20). Here, using a digital planimetry method, we examined the high-resolution segment image sets of both MRI and histological sections. The planimetry method is a reliable method for volume estimation in which areas of interest are outlined on a computer screen and the included pixels in series of images are used for volume calculation (28). Using this method, we demonstrated a highly significant correlation between the hippocampal volume determined from the histological sections and that determined using MRI, showing that MRI can provide accurate relative differences over time.
In addition to developing MRI methods for estimating brain volume, by examining 2- and 5-wk-old piglets, this study also provides evidence of substantial brain growth during this early 3-wk period. A limitation of this study was that it was not a longitudinal design due to the need to obtain brain sections for the estimation of hippocampal volume using the Cavalieri method. Nonetheless, the volumes for cortex and subcortical regions including the hippocampus, diencephalon, cerebellum, and brainstem were 30–43% greater in 5-wk-old piglets as compared with 2-wk-old piglets. No main effect of sex on absolute volumes or differences in volumes between 2- and 5-wk-old piglets was evident for these brain regions. These data are similar to a study of humans aged 3 mo to 30 y that found no effects of sex on gray and white matter volumes after accounting for head size (15).
Additional limitations of this study include the simplicity of the volumetric analysis technique and the histological methods used for the validation of the MRI volumes. Due to the lack of age-specific MRI-based atlases for the neonatal piglet, more sophisticated regional volume estimation methods could not be used. Our lab is collecting MRI data to construct these averaged brain MRI atlases to utilize techniques such as voxel- and deformation-based morphology, which have been shown to be powerful methods for volumetric analysis in the neonatal brain (29). In addition, the automated skull-stripping features of many semi- and fully automated procedures that were developed for humans do not currently work with the pig due to anatomical differences. For this study, only two independent observers were evaluated to construct the coefficients of variation. Although it would be ideal to evaluate additional observers, we obtained low coefficients of variation for our methods, and the use of only two observers is similar to other manual segmentation protocols for the analysis of human brain regions (30,31). For the histological validation, there were differences in the absolute volumes between the MRI and histologically determined hippocampal volumes. These differences were due to the dehydration and paraffin-embedding of the hippocampal tissue. We chose this approach in order to be able to slice the tissue at smaller intervals to increase the resolution of our histological method. Because of this, the correlation analysis is based on relative differences in volume rather than absolute volumes. In addition, another limitation is that tissues from only four brains were used for the correlation analysis, although all of these brains were from littermate pigs, allowing for reduction in variability. Despite the limitations of this study, the data support this as a reliable method for quantification of brain region volumes in the neonatal piglet, and future work will allow for more semi- and fully automated segmentation of the piglet brain.
In summary, reliable estimates of changes in the brain volume over time in neonatal piglets can be obtained using MRI and manual segmentation procedures. Because the pig is a gyrencephalic species thought to have brain growth and development similar to humans, it may serve as a preclinical translational model for studying the developmental origins of neuropsychiatric diseases. Using the methods introduced in this study, ongoing studies will characterize normal brain development in the pig from the neonatal period to adulthood in a longitudinal design.
Methods
Subjects
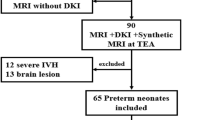
A total of 12 pigs, six male and six female (Sus scrofa domestica, York breed), from three litters, were obtained from the University of Illinois swine herd at 2 d of age and placed into an artificial rearing system that has been previously described (9). Briefly, each piglet was housed individually in an acrylic-sided cage with a 12-h light/dark cycle. Ambient temperature was maintained at 27 °C with intracage temperatures maintained at 32 °C using radiant heaters located directly over the piglets. Piglets received a commercially available milk replacer (Milk Specialties Global Animal Nutrition, Carpentersville, IL) throughout the 5-wk study. Milk replacer powder was reconstituted fresh each day and 50 ml of liquid diet was delivered to individual feeding bowls once an hour from 8 AM to midnight using a custom, automated system (9). The pigs had an average weight of 1.86 ± 0.05 kg at birth and 3.08 ± 0.05 kg and 9.13 ± 0.34 kg at 2 and 5 wks of age, respectively.
Experimental Procedures
At 2 wks of age, six of the pigs, three male and three female, were subjected to MRI procedures. Immediately before scanning, pigs were anesthetized by intramuscular injection of a telazol:ketamine:xylazine solution (4.4 mg/kg body weight), and placed in a prone position in the MRI scanner. The pigs remained anesthetized during the MRI scanning procedure (~20 min) and were later killed by intracardiac administration of an overdose of sodium pentobarbital (390 mg/ml Fatal Plus—administered at a rate of 1 ml/5 kg body weight). Brains were removed and a 2.5-cm3 tissue block encompassing the left hippocampus was excised and placed in 4% paraformaldehyde. Tissue blocks were stored in the fixative at 4 °C for 3 wks before processing. Tissue blocks from male and female littermates were selected and processed for the determination of histological volume. The remaining six pigs were scanned at 5 wks of age after which they were killed and their brain tissue was collected and processed as described above.
Imaging
Magnetic resonance scanning was conducted using a Siemens MAGNETOM Trio 3T imager by employing a Siemens 32-channel head coil (Siemens, Erlangen, Germany). Anatomic images were acquired using a 3D T1-weighted magnetization-prepared rapid gradient-echo sequence with the following parameters: repetition time = 1,900 ms; echo time = 2.48 ms; inversion time = 900 ms, flip angle = 9°, matrix = 256 × 256 (interpolated to 512 × 512), slice thickness = 1.0 mm. The final voxel size was 0.35 × 0.35 × 1.0 mm across the entire head from the tip of the snout to the cervical/thoracic spinal cord junction. Images were exported in the Digital Imaging and Communications in Medicine format and analyzed using 3D visualization software (AMIRA; Visage Imaging, San Diego, CA).
In vivo Quantification of Brain Region Volume Using MRI
The volumes for total brain, cortex, diencephalon, cerebellum, hippocampus, and brainstem were quantified using AMIRA 3D visualization software. Unsharp masking post-processing was performed to improve the grayscale inhomogeneities (32). Manual segmentation of the brain regions was facilitated by a Wacom Cintiq 21UX graphic input screen and pen (Wacom, Vancouver, WA). The anatomical criteria for the regions were based on the stereotaxic atlas of Félix et al. (33). The boundaries for segmentation are shown in Figure 2 . For the hippocampus, the medial border was defined by the choroidal fissure and the lateral border was the inferior horn of the lateral ventricle. The superior border was the body of the lateral ventricle and the inferior border was the parahippocampal gyrus. The total brain volume was defined as the sum of cortical, subcortical, cerebellar, brainstem, and partial spinal cord volumes. The spinal cord was segmented to the posterior boundary of the cerebellum. The volumes were calculated using the material statistics function of AMIRA, which estimates the 3D volume of the manually segmented regions given the aforementioned voxel size. To determine the reliability of these measures, segmentation was conducted by two independent observers and the intra- and interobserver agreements calculated.
Quantification of Hippocampal Volume from Histological Sections
Paraformaldehyde-fixed hippocampal tissue was dehydrated and paraffin-embedded using a Leica ASP300 tissue processor (Leica, Wetzlar, Germany). The tissue block was sectioned in the axial plane at 5-µm intervals and every 10th section was collected. The serial sections were hematoxylin and eosin–stained using a Sakura Tissue-Tek DRS H&E stainer system and coverslipped with a Sakura Tissue-Tek Glas coverslipper (Sakura, Tokyo, Japan). The slides were digitized at ×20 magnification using a Nanozoomer Digital Pathology System (Hamamatsu, Hamamatsu City, Japan), creating high-resolution virtual slide images. Digital images of the hippocampal region were exported from the virtual slides as JPEGs for subsequent segmentation. Digital images of the serial sections were imported into AMIRA, and the z-plane depth was adjusted to account for the 50-µm distance between the collected slices. Images were aligned and the hippocampus was manually segmented for each section. The hippocampal region was defined as the dentate gyrus, CA1, and CA3, as shown in a representative slide in Figure 3 . After the segmentation of all tissue sections, the material statistics function of AMIRA was used to estimate the hippocampal volume.
Hippocampus segmentation criteria for histological tissue sections. The outlined area includes the dentate gyrus (DG), CA1, and CA3 regions of a horizontal cross-sectional slice of the hippocampal tissue from the dorsobasal portion of the hippocampus. The subiculum (Sub) was not included in volume analysis. Image was taken at ×12.5 original magnification.
Statistics
For the analysis of the brain region volumes in the 2- and 5-wk-old piglets, data were subjected to two-way ANOVA (age × sex) using SAS software (SAS, Cary, NC). It was determined that there was unequal variance between the data obtained for 2- and 5-wk-old piglets, and to correct for this, the data were log-transformed before ANOVA. There was no significant effect of sex; therefore, the male and female data were combined for the calculation of the means. Significance level was set at P < 0.05. Validation of the MRI volume estimation of the hippocampus was based on the technique employed in human studies by Bobinski et al. (34). The correlative relationship between the hippocampal volumes obtained by MRI and histological measurement was determined using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). The Pearson product–moment correlation coefficient was used to determine the strength of the linear dependence between the two volume estimations.
The intra- and interobserver agreements for manual segmentation were calculated based on the statistical methods by Bland and Altman (35). The intraobserver agreement was calculated by the following formula, where x1st and x2nd were repeat measures of the regional volumes of six pigs using manual segmentation by one rater (M.S.C). This rater was blinded to the pig brain images being segmented and the order of segmentation was randomized.

The interobserver agreement was calculated by the following formula where xa and xb are measures of regional volumes of each pig using manual segmentation by two different raters (M.S.C and A.N.).

The intraobserver coefficient of variation was defined as 2 SD of the following equation by the British Standards Institution (36):

The interobserver coefficient of variation was defined as 2 SD of the following equation:

Ethics
All procedures were in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and approved by the University of Illinois Institutional Animal Care and Use Committee.
References
Dixon JA, Spinale FG. Large animal models of heart failure: a critical link in the translation of basic science to clinical practice. Circ Heart Fail 2009;2:262–71.
Bellinger DA, Merricks EP, Nichols TC . Swine models of type 2 diabetes mellitus: insulin resistance, glucose tolerance, and cardiovascular complications. ILAR J 2006;47:243–58.
Miller ER, Ullrey DE . The pig as a model for human nutrition. Annu Rev Nutr 1987;7:361–82.
Lind NM, Moustgaard A, Jelsing J, Vajta G, Cumming P, Hansen AK . The use of pigs in neuroscience: modeling brain disorders. Neurosci Biobehav Rev 2007;31:728–51.
Pond WG, Boleman SL, Fiorotto ML, et al. Perinatal ontogeny of brain growth in the domestic pig. Proc Soc Exp Biol Med 2000;223:102–8.
Dobbing J, Sands J . Comparative aspects of the brain growth spurt. Early Hum Dev 1979;3:79–83.
Dickerson JWT, Dobbing J . Prenatal and postnatal growth and development of the central nervous system of the pig. Proc R Soc Lond 1967;166:384–95.
Thibault KL, Margulies SS . Age-dependent material properties of the porcine cerebrum: effect on pediatric inertial head injury criteria. J Biomech 1998;31:1119–26.
Dilger RN, Johnson RW . Behavioral assessment of cognitive function using a translational neonatal piglet model. Brain Behav Immun 2010;24:1156–65.
Ganessunker D, Gaskins HR, Zuckermann FA, Donovan SM . Total parenteral nutrition alters molecular and cellular indices of intestinal inflammation in neonatal piglets. JPEN J Parenter Enteral Nutr 1999;23:337–44.
Wood NS, Marlow N, Costeloe K, Gibson AT, Wilkinson AR . Neurologic and developmental disability after extremely preterm birth. EPICure Study Group. N Engl J Med 2000;343:378–84.
Giedd JN, Vaituzis AC, Hamburger SD,et al. Quantitative MRI of the temporal lobe, amygdala, and hippocampus in normal human development: ages 4-18 years. J Comp Neurol 1996;366:223–30.
Knickmeyer RC, Gouttard S, Kang C,et al. A structural MRI study of human brain development from birth to 2 years. J Neurosci 2008;28:12176–82.
Giedd JN, Blumenthal J, Jeffries NO,et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci 1999;2:861–3.
Pfefferbaum A, Mathalon DH, Sullivan EV, Rawles JM, Zipursky RB, Lim KO . A quantitative magnetic resonance imaging study of changes in brain morphology from infancy to late adulthood. Arch Neurol 1994;51:874–87.
Gilmore JH, Lin W, Prastawa MW,et al. Regional gray matter growth, sexual dimorphism, and cerebral asymmetry in the neonatal brain. J Neurosci 2007;27:1255–60.
Dubois J, Benders M, Cachia A,et al. Mapping the early cortical folding process in the preterm newborn brain. Cereb Cortex 2008;18:1444–54.
Thompson DK, Wood SJ, Doyle LW, et al. Neonate hippocampal volumes: prematurity, perinatal predictors, and 2-year outcome. Ann Neurol 2008;63:642–51.
Beauchamp MH, Thompson DK, Howard K, et al. Preterm infant hippocampal volumes correlate with later working memory deficits. Brain 2008;131(Pt 11):2986–94.
Jelsing J, Rostrup E, Markenroth K, et al. Assessment of in vivo MR imaging compared to physical sections in vitro–a quantitative study of brain volumes using stereology. Neuroimage 2005;26:57–65.
Kuluz J, Samdani A, Benglis D, et al. Pediatric spinal cord injury in infant piglets: description of a new large animal model and review of the literature. J Spinal Cord Med 2010;33:43–57.
Rosendal F, Pedersen M, Sangill R, et al. MRI protocol for in vivo visualization of the Göttingen minipig brain improves targeting in experimental functional neurosurgery. Brain Res Bull 2009;79:41–5.
Sandberg DI, Crandall KM, Koru-Sengul T, et al. Pharmacokinetic analysis of etoposide distribution after administration directly into the fourth ventricle in a piglet model. J Neurooncol 2010;97:25–32.
Björkman ST, Miller SM, Rose SE, Burke C, Colditz PB . Seizures are associated with brain injury severity in a neonatal model of hypoxia-ischemia. Neuroscience 2010;166:157–67.
Munkeby BH, De Lange C, Emblem KE, et al. A piglet model for detection of hypoxic-ischemic brain injury with magnetic resonance imaging. Acta Radiol 2008;49:1049–57.
Shi F, Fan Y, Tang S, Gilmore JH, Lin W, Shen D . Neonatal brain image segmentation in longitudinal MRI studies. Neuroimage 2010;49:391–400.
Saikali S, Meurice P, Sauleau P, et al. A three-dimensional digital segmented and deformable brain atlas of the domestic pig. J Neurosci Methods 2010;192:102–9.
Cotter D, Miszkiel K, Al-Sarraj S, et al. The assessment of postmortem brain volume; a comparison of stereological and planimetric methodologies. Neuroradiology 1999;41:493–6.
Boardman JP, Counsell SJ, Rueckert D, et al. Abnormal deep grey matter development following preterm birth detected using deformation-based morphometry. Neuroimage 2006;32:70–8.
Bokde AL, Teipel SJ, Schwarz R, et al. Reliable manual segmentation of the frontal, parietal, temporal, and occipital lobes on magnetic resonance images of healthy subjects. Brain Res Brain Res Protoc 2005;14:135–45.
Yu X, Zhang Y, Lasky RE, Datta S, Parikh NA, Narayana PA . Comprehensive brain MRI segmentation in high risk preterm newborns. PLoS ONE 2010;5:e13874.
Brinkmann BH, Manduca A, Robb RA . Optimized homomorphic unsharp masking for MR grayscale inhomogeneity correction. IEEE Trans Med Imaging 1998;17:161–71.
Félix B, Léger ME, Albe-Fessard D, Marcilloux JC, Rampin O, Laplace JP . Stereotaxic atlas of the pig brain. Brain Res Bull 1999;49:1–137.
Bobinski M, de Leon MJ, Wegiel J, et al. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer’s disease. Neuroscience 2000;95:721–5.
Bland JM, Altman DG . Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986;1:307–10.
British Standards Institution 1998 Accuracy (trueness and precision) of measurement methods and results. Alternative methods for the determination of the precision of a standard measurement method. BS ISO 5725-5:1998.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Conrad, M., Dilger, R., Nickolls, A. et al. Magnetic resonance imaging of the neonatal piglet brain. Pediatr Res 71, 179–184 (2012). https://doi.org/10.1038/pr.2011.21
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2011.21
This article is cited by
-
A novel model of acquired hydrocephalus for evaluation of neurosurgical treatments
Fluids and Barriers of the CNS (2021)
-
Cerebral organoids and their potential for studies of brain diseases in domestic animals
Veterinary Research (2021)
-
Comparing interspecific socio-communicative skills of socialized juvenile dogs and miniature pigs
Animal Cognition (2019)