Abstract
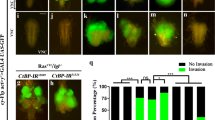
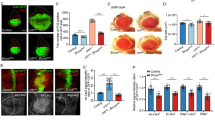
Autophagy is a catabolic process that has been implicated both as a tumor suppressor and in tumor progression. Here, we investigate this dichotomy in cancer biology by studying the influence of altered autophagy in Drosophila models of tissue overgrowth. We find that the impact of altered autophagy depends on both genotype and cell type. As previously observed in mammals, decreased autophagy suppresses Ras-induced eye epithelial overgrowth. In contrast, autophagy restricts epithelial overgrowth in a Notch-dependent eye model. Even though decreased autophagy did not influence Hippo pathway-triggered overgrowth, activation of autophagy strongly suppresses this eye epithelial overgrowth. Surprisingly, activation of autophagy enhanced Hippo pathway-driven overgrowth in glia cells. These results indicate that autophagy has different influences on tissue growth in distinct contexts, and highlight the importance of understanding the influence of autophagy on growth to augment a rationale therapeutic strategy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kondo Y, Kanzawa T, Sawaya R, Kondo S . The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 2005; 5: 726–734.
Mizushima N, Komatsu M . Autophagy: renovation of cells and tissues. Cell 2011; 147: 728–741.
Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007; 21: 1367–1381.
Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21: 1621–1635.
Mathew R, Karantza-Wadsworth V, White E . Role of autophagy in cancer. Nat Rev Cancer 2007; 7: 961–967.
Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013; 504: 296–300.
Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014; 5: 3056.
Funderburk SF, Wang QJ, Yue Z . The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20: 355–362.
Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH . Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development 2013; 140: 1321–1329.
Tipping M, Perrimon N . Drosophila as a model for context-dependent tumorigenesis. J Cell Physiol 2014; 229: 27–33.
Rudrapatna VA, Cagan RL, Das TK . Drosophila cancer models. Dev Dyn 2012; 241: 107–118.
Bos JL . ras oncogenes in human cancer: a review. Cancer Res 1989; 49: 4682–4689.
Karim FD, Rubin GM . Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 1998; 125: 1–9.
Bergmann A, Agapite J, McCall K, Steller H . The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 1998; 95: 331–341.
Kurada P, White K . Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 1998; 95: 319–329.
Lobry C, Oh P, Aifantis I . Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med 2011; 208: 1931–1935.
Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Aviño FJ, Dominguez M . Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 2006; 439: 430–436.
Johnson R, Halder G . The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov 2014; 13: 63–79.
Elsum I, Yates L, Humbert PO, Richardson HE . The Scribble-Dlg-Lgl polarity module in development and cancer: from flies to man. Essays Biochem 2012; 53: 141–168.
Pagliarini RA, Xu T . A genetic screen in Drosophila for metastatic behavior. Science 2003; 302: 1227–1231.
Igaki T, Pagliarini RA, Xu T . Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol 2006; 16: 1139–1146.
Brumby AM, Richardson HE . scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J 2003; 22: 5769–5779.
Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 2013; 27: 1447–1461.
Scott RC, Juhász G, Neufeld TP . Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 2007; 17: 1–11.
Berry DL, Baehrecke EH . Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 2007; 131: 1137–1148.
Chang T-K, Shravage BV, Hayes SD, Powers CM, Simin RT, Harper JW et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol 2013; 15: 1067–1078.
Reddy BV, Irvine KD . Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development 2011; 138: 5201–5212.
Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25: 795–800.
Vander Heiden MG, Cantley LC, Thompson CB . Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033.
McPhee CK, Baehrecke EH . Autophagy in Drosophila melanogaster. Biochim Biophys Acta 2009; 1793: 1452–1460.
Scott RC, Schuldiner O, Neufeld TP . Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7: 167–178.
Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J et al. Autophagy termination and lysosome reformation regulated by mTOR. Nature 2010; 465: 942–946.
Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM . Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141: 290–303.
Dutta S, Baehrecke EH . Warts is required for PI3K-regulated growth arrest, autophagy, and autophagic cell death in Drosophila. Curr Biol 2008; 18: 1466–1475.
Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 2011; 25: 460–470.
Huang J, Wu S, Barrera J, Matthews K, Pan D . The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005; 122: 421–434.
Chen CL, Schroeder MC, Kango-Singh M, Tao C, Halder G . Tumor suppression by cell competition through regulation of the Hippo pathway. Proc Natl Acad Sci USA 2012; 109: 484–489.
Chang YY, Neufeld TP . An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 2009; 20: 2004–2014.
Brand AH, Perrimon N . Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993; 118: 401–415.
Lee T, Luo L . Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 2001; 24: 251–254.
Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K et al. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS ONE 2012; 7: e44214.
Acknowledgements
We thank T Chang, M Dominguez, M Freeman, G Halder, G Juhasz, M Kango-Singh, TP Neufeld, the Bloomington Stock Center, the VDRC for flies and antibodies, J Lindblad, C Nelson and T Fortier for technical support, and the Bergmann and Baehrecke labs for constructive comments. This work was supported by NIH grants GM068016 to AB and CA159314 to EHB. EHB is an Ellison Medical Foundation Scholar.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Pérez, E., Das, G., Bergmann, A. et al. Autophagy regulates tissue overgrowth in a context-dependent manner. Oncogene 34, 3369–3376 (2015). https://doi.org/10.1038/onc.2014.285
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.285
This article is cited by
-
Innate immune and proinflammatory signals activate the Hippo pathway via a Tak1-STRIPAK-Tao axis
Nature Communications (2024)
-
Hippo pathway effectors YAP and TAZ and their association with skeletal muscle ageing
Journal of Physiology and Biochemistry (2021)
-
Emerging role of the Hippo pathway in autophagy
Cell Death & Disease (2020)
-
Colorectal cancer cells respond differentially to autophagy inhibition in vivo
Scientific Reports (2019)
-
Microenvironmental autophagy promotes tumour growth
Nature (2017)