Abstract
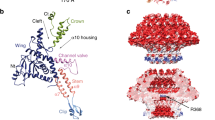
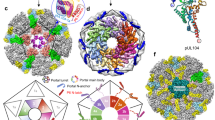
DNA viruses such as bacteriophages and herpesviruses deliver their genome into and out of the capsid through large proteinaceous assemblies, known as portal proteins. Here, we report two snapshots of the dodecameric portal protein of bacteriophage P22. The 3.25-Å-resolution structure of the portal-protein core bound to 12 copies of gene product 4 (gp4) reveals a ~1.1-MDa assembly formed by 24 proteins. Unexpectedly, a lower-resolution structure of the full-length portal protein unveils the unique topology of the C-terminal domain, which forms a ~200-Å-long α-helical barrel. This domain inserts deeply into the virion and is highly conserved in the Podoviridae family. We propose that the barrel domain facilitates genome spooling onto the interior surface of the capsid during genome packaging and, in analogy to a rifle barrel, increases the accuracy of genome ejection into the host cell.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lander, G.C. et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312, 1791–1795 (2006).
Johnson, J.E. & Chiu, W. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 17, 237–243 (2007).
Casjens, S. & Weigele, P. Headful DNA packaging by bacteriophage P22. in Viral Genome Packaging Machines: Genetics, Structure and Mechanism (ed. Catalano, C.) 80–88 (Landes Publishing, Georgetown, Texas, USA, 2005).
Ackermann, H.W. Bacteriophage observations and evolution. Res. Microbiol. 154, 245–251 (2003).
Teschke, C.M. & Parent, K.N. 'Let the phage do the work': using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 401, 119–130 (2010).
Chang, J., Weigele, P., King, J., Chiu, W. & Jiang, W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14, 1073–1082 (2006).
Lander, G.C. et al. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure 17, 789–799 (2009).
Olia, A.S., Bhardwaj, A., Joss, L., Casjens, S. & Cingolani, G. Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry 46, 8776–8784 (2007).
Strauss, H. & King, J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol. 172, 523–543 (1984).
Olia, A.S., Casjens, S. & Cingolani, G. Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas. Protein Sci. 18, 537–548 (2009).
Olia, A.S., Casjens, S. & Cingolani, G. Structure of phage P22 cell envelope-penetrating needle. Nat. Struct. Mol. Biol. 14, 1221–1226 (2007).
Bhardwaj, A., Walker-Kopp, N., Casjens, S.R. & Cingolani, G. An evolutionarily conserved family of virion tail needles related to bacteriophage P22 gp26: correlation between structural stability and length of the alpha-helical trimeric coiled coil. J. Mol. Biol. 391, 227–245 (2009).
Olia, A.S. et al. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J. Mol. Biol. 363, 558–576 (2006).
Lorenzen, K., Olia, A.S., Uetrecht, C., Cingolani, G. & Heck, A.J. Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J. Mol. Biol. 379, 385–396 (2008).
Israel, V. E proteins of bacteriophage P22. I. Identification and ejection from wild-type and defective particles. J. Virol. 23, 91–97 (1977).
Lenk, E., Casjens, S., Weeks, J. & King, J. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology 68, 182–199 (1975).
Simpson, A.A. et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408, 745–750 (2000).
Bazinet, C., Benbasat, J., King, J., Carazo, J.M. & Carrascosa, J.L. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry 27, 1849–1856 (1988).
Simpson, A.A. et al. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr. D Biol. Crystallogr. 57, 1260–1269 (2001).
Guasch, A. et al. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 315, 663–676 (2002).
Guo, P.X., Erickson, S. & Anderson, D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science 236, 690–694 (1987).
Orlova, E.V. et al. Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol. 6, 842–846 (1999).
Lebedev, A.A. et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 26, 1984–1994 (2007).
Lurz, R. et al. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 310, 1027–1037 (2001).
Orlova, E.V. et al. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 22, 1255–1262 (2003).
Cingolani, G., Moore, S.D., Prevelige, P.E. Jr. & Johnson, J.E. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J. Struct. Biol. 139, 46–54 (2002).
Trus, B.L. et al. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78, 12668–12671 (2004).
Newcomb, W.W. et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75, 10923–10932 (2001).
Zheng, H. et al. A conformational switch in bacteriophage p22 portal protein primes genome injection. Mol. Cell 29, 376–383 (2008).
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B. & Hughes, C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919 (2000).
Steinbacher, S. et al. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265, 383–386 (1994).
Steinbacher, S. et al. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267, 865–880 (1997).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Moore, S.D. & Prevelige, P.E. Jr. Structural transformations accompanying the assembly of bacteriophage P22 portal protein rings in vitro. J. Biol. Chem. 276, 6779–6788 (2001).
Guo, L., Han, A., Bates, D.L., Cao, J. & Chen, L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc. Natl. Acad. Sci. USA 104, 4297–4302 (2007).
Eppler, K., Wyckoff, E., Goates, J., Parr, R. & Casjens, S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology 183, 519–538 (1991).
Lhuillier, S. et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl. Acad. Sci. USA 106, 8507–8512 (2009).
Cardarelli, L. et al. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J. Mol. Biol. 395, 754–768 (2010).
Zhao, H. et al. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc. Natl. Acad. Sci. USA 107, 1971–1976 (2010).
Vlieghe, D., Turkenburg, J.P. & Van Meervelt, L. B-DNA at atomic resolution reveals extended hydration patterns. Acta Crystallogr. D Biol. Crystallogr. 55, 1495–1502 (1999).
Hendrix, R.W. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. USA 75, 4779–4783 (1978).
Casjens, S. et al. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 224, 1055–1074 (1992).
Cuervo, A., Vaney, M.C., Antson, A.A., Tavares, P. & Oliveira, L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J. Biol. Chem. 282, 18907–18913 (2007).
Hugel, T. et al. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 5, e59 (2007).
Baumann, R.G., Mullaney, J. & Black, L.W. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 61, 16–32 (2006).
Guy, A.E. A practical method of calculating the powder pressure and the velocity of projectile along the bore of a cannon. J. Am. Soc. Nav. Eng. 33, 720–734 (1921).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Collaborative Computational Project. Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Painter, J. & Merritt, E.A. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 (2006).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank V. Stojanoff and staff at the National Synchrotron Light Source beamlines X6A, X25 and X29A and the staff of the Macromolecular Diffraction Facility at the Cornell High Energy Synchrotron Source (macCHESS) for beam time and assistance in data collection. This work was supported by US National Institutes of Health grants 1R56 AI076509-01A1 (to G.C.) and RO1 AI40101 (to J.E.J.).
Author information
Authors and Affiliations
Contributions
A.S.O. and G.C. crystallized the full-length portal and portal-protein core–gp4 complex, collected the X-ray data and determined the structures. P.E.P. isolated the gene encoding P22 portal protein and helped with data analysis. J.E.J. supervised the crystallization and data collection of full-length portal protein and helped with data analysis. G.C. coordinated the overall project and wrote the manuscript with A.S.O.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 4149 kb)
Rights and permissions
About this article
Cite this article
Olia, A., Prevelige, P., Johnson, J. et al. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol 18, 597–603 (2011). https://doi.org/10.1038/nsmb.2023
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2023
This article is cited by
-
Structure and proposed DNA delivery mechanism of a marine roseophage
Nature Communications (2023)
-
Phage diversity, genomics and phylogeny
Nature Reviews Microbiology (2020)
-
Structural changes of a bacteriophage upon DNA packaging and maturation
Protein & Cell (2020)
-
Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism
Nature Communications (2019)
-
Genomic and biological characterization of the Vibrio alginolyticus-infecting “Podoviridae” bacteriophage, vB_ValP_IME271
Virus Genes (2019)