Abstract
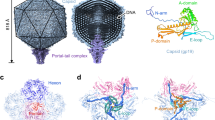
Podovirus P-SSP7 infects Prochlorococcus marinus, the most abundant oceanic photosynthetic microorganism. Single-particle cryo–electron microscopy yields icosahedral and asymmetrical structures of infectious P-SSP7 with 4.6-Å and 9-Å resolution, respectively. The asymmetric reconstruction reveals how symmetry mismatches are accommodated among five of the gene products at the portal vertex. Reconstructions of infectious and empty particles show a conformational change of the 'valve' density in the nozzle, an orientation difference in the tail fibers, a disordering of the C terminus of the portal protein and the disappearance of the core proteins. In addition, cryo–electron tomography of P-SSP7 infecting Prochlorococcus showed the same tail-fiber conformation as that in empty particles. Our observations suggest a mechanism whereby, upon binding to the host cell, the tail fibers induce a cascade of structural alterations of the portal vertex complex that triggers DNA release.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sullivan, M.B., Coleman, M.L., Weigele, P., Rohwer, F. & Chisholm, S.W. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3, e144 (2005).
Moore, L.R., Rocap, G. & Chisholm, S.W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467 (1998).
Liu, H., Nolla, H.A. & Campbell, L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical north Pacific Ocean. Aquat. Microb. Ecol. 12, 39–47 (1997).
Angly, F.E. et al. The marine viromes of four oceanic regions. PLoS Biol. 4, e368 (2006).
Lindell, D. et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449, 83–86 (2007).
Lindell, D., Jaffe, J.D., Johnson, Z.I., Church, G.M. & Chisholm, S.W. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438, 86–89 (2005).
Liu, X., Jiang, W., Jakana, J. & Chiu, W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a multi-path simulated annealing optimization algorithm. J. Struct. Biol. 160, 11–27 (2007).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Jiang, W. et al. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature 451, 1130–1134 (2008).
Wikoff, W.R. et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289, 2129–2133 (2000).
Jiang, W. et al. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 10, 131–135 (2003).
Fokine, A. et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. USA 102, 7163–7168 (2005).
Morais, M.C. et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of φ29. Mol. Cell 18, 149–159 (2005).
Baker, M.L. et al. Architecture of the herpes simplex virus major capsid protein derived from structural bioinformatics. J. Mol. Biol. 331, 447–456 (2003).
Purohit, P.K. et al. Forces during bacteriophage DNA packaging and ejection. Biophys. J. 88, 851–866 (2005).
Aksyuk, A.A. et al. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 28, 821–829 (2009).
Orlova, E.V. et al. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 22, 1255–1262 (2003).
Lhuillier, S. et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl. Acad. Sci. USA 106, 8507–8512 (2009).
Leiman, P.G., Chipman, P.R., Kostyuchenko, V.A., Mesyanzhinov, V.V. & Rossmann, M.G. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118, 419–429 (2004).
Plisson, C. et al. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 26, 3720–3728 (2007).
Cong, Y. et al. Structural mechanism of SDS-induced enzyme activity of scorpion hemocyanin revealed by electron cryomicroscopy. Structure 17, 749–758 (2009).
Henderson, R. & Moffat, J.K. The difference Fourier technique in protein crystallography: errors and their treatment. Acta Crystallogr. B 27, 1414–1420 (1971).
Kemp, P., Garcia, L.R. & Molineux, I.J. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 340, 307–317 (2005).
Baker, M.L., Ju, T. & Chiu, W. Identification of secondary structure elements in intermediate-resolution density maps. Structure 15, 7–19 (2007).
Simpson, A.A. et al. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr. D Biol. Crystallogr. 57, 1260–1269 (2001).
Lebedev, A.A. et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 26, 1984–1994 (2007).
McGuffin, L.J., Bryson, K. & Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 (2000).
Cole, C., Barber, J.D. & Barton, G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 (2008).
Guo, L., Han, A., Bates, D.L., Cao, J. & Chen, L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc. Natl. Acad. Sci. USA 104, 4297–4302 (2007).
Jiang, W. et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439, 612–616 (2006).
Randall, G.L., Zechiedrich, L. & Pettitt, B.M. In the absence of writhe, DNA relieves torsional stress with localized, sequence-dependent structural failure to preserve B-form. Nucleic Acids Res. 37, 5568–5577 (2009).
Tang, J. et al. DNA poised for release in bacteriophage φ29. Structure 16, 935–943 (2008).
Hendrix, R.W. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. USA 75, 4779–4783 (1978).
Simpson, A.A. et al. Structure of the bacteriophage φ29 DNA packaging motor. Nature 408, 745–750 (2000).
Agirrezabala, X. et al. Maturation of phage T7 involves structural modi-fication of both shell and inner core components. EMBO J. 24, 3820–3829 (2005).
Kostyuchenko, V.A. et al. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 12, 810–813 (2005).
Yu, X., Jin, L. & Zhou, Z.H. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453, 415–419 (2008).
Lander, G.C. et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312, 1791–1795 (2006).
Leiman, P.G. et al. The structures of bacteriophages K1E and K1–5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J. Mol. Biol. 371, 836–849 (2007).
Xiang, Y. et al. Structural changes of bacteriophage φ29 upon DNA packaging and release. EMBO J. 25, 5229–5239 (2006).
Serysheva, I.I. et al. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl. Acad. Sci. USA 105, 9610–9615 (2008).
Baker, M.L., Marsh, M.P. & Chiu, W. Cryo-EM of molecular nanomachines and cells. in Nanotechnology Vol. 5 (ed. Vogel, V.) 91–111 (Wiley-VCH, Hoboken, New Jersey, USA, 2009).
Lucic, V., Forster, F. & Baumeister, W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74, 833–865 (2005).
Steven, A.C. et al. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200, 351–365 (1988).
Molineux, I.J. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40, 1–8 (2001).
Molineux, I.J., Calendar, R. & Abedon, S.T. The T7 group. in The Bacteriophages Vol. 17 (Oxford University Press USA, New York, USA, 2006).
Liu, X., Rochat, R.H & Chiu, W. Reconstructing cyano-bacteriophage P-SSP7 structure without imposing symmetry. Nat. Protoc. published online, doi:10.1038/nprot.2010.96 (13 June 2010).
Kivioja, T., Ravantti, J., Verkhovsky, A., Ukkonen, E. & Bamford, D. Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 131, 126–134 (2000).
Crowther, R.A., DeRosier, D.J. & Klug, A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc. R. Soc. Lond. A Math. Phys. Sci. 317, 319–340 (1970).
Chang, J., Weigele, P., King, J., Chiu, W. & Jiang, W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14, 1073–1082 (2006).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Ludtke, S.J. et al. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure 16, 441–448 (2008).
Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Kremer, J.R., Mastronarde, D.N. & McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Schmid, M.F. & Booth, C.R. Methods for aligning and for averaging 3D volumes with missing data. J. Struct. Biol. 161, 243–248 (2008).
Acknowledgements
We thank M. Coleman, S. Rodrigue, R. Malmstrom, J. Waldbauer and S. Kern for assistance in the infection experiments, J. Chang, Q. Zeng, S. Labrie and F. Rixon for discussions and J. Chang, K. Welgehausen and R. Rochat for editorial assistance in preparation of the manuscript. This research was supported in parts by grants from the US National Institutes of Health (P41RR002250, R01GM079429 to W.C.), the US National Science Foundation (IIS-0705644 to W.C. and M.L.B.), the Robert Welch Foundation (Q1242 to W.C.), the US Department of Energy, the US National Science Foundation and the Gordon and Betty Moore Foundation (to S.W.C.).
Author information
Authors and Affiliations
Contributions
M.B.S., M.S.O. and S.W.C. designed and conducted the phage-host experiments and provided the samples; Q.Z. took single-particle images; Q.Z., C.F. and K.M. took tomographic images; X.L. introduced the methodology for and performed the single-particle symmetric and asymmetric reconstructions; X.L. and W.C. interpreted the maps; M.F.S. analyzed the difference maps; K.M. did tomographic reconstructions and performed analysis with assistance from M.F.S.; M.L.B. and X.L. built Cα backbone models; M.T.D. and W.C. designed the animations; X.L. and W.C. wrote the paper with contributions from all coauthors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Figures 1–8 (PDF 3879 kb)
Supplementary Video 1
4.6 Å capsid density and backbone models derived from the map with icosahedral symmetry imposed. (a) density map and segmented 7 asymmetric subunits. (MOV 2232 kb)
Supplementary Video 2
Backbone model for 1 subunit. (MOV 1780 kb)
Supplementary Video 3
Backbone models for the whole capsid. (MOV 2945 kb)
Supplementary Video 4
9 Å density map of P-SSP7 phage with no symmetry imposed.Different components comprising the portal vertex machinery are annotated as nozzle, adaptor, portal, tail fiber, core protein and DNA terminus. (MOV 2336 kb)
Supplementary Video 5
Tomogram of P-SSP7 infecting a Prochlorococcus (MED4 strain) cell is shown along the z-direction. (MOV 2840 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Zhang, Q., Murata, K. et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol 17, 830–836 (2010). https://doi.org/10.1038/nsmb.1823
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1823
This article is cited by
-
Abundant and cosmopolitan lineage of cyanopodoviruses lacking a DNA polymerase gene
The ISME Journal (2023)
-
Structure and proposed DNA delivery mechanism of a marine roseophage
Nature Communications (2023)
-
Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism
Nature Communications (2019)
-
Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation
Nature Communications (2017)
-
Genetic hurdles limit the arms race between Prochlorococcus and the T7-like podoviruses infecting them
The ISME Journal (2017)