Abstract
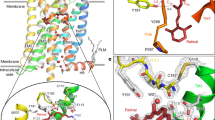
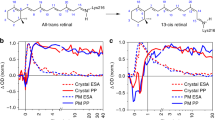
Protein photoreceptors use small-molecule cofactors called chromophores to detect light. Only under the influence of the receptors' active sites do these chromophores adopt spectral and photochemical properties that suit the receptors' functional requirements. This protein-induced change in chromophore properties is called photochemical tuning and is a prime example for the general—but poorly understood—process of chemical tuning through which proteins shape the reactivity of their active-site groups. Here we report the 0.82-Å resolution X-ray structure of the bacterial light receptor photoactive yellow protein (PYP). The unusually precise structure reveals deviations from expected molecular geometries and anisotropic atomic displacements in the PYP active site. Our analysis of these deviations points directly to the intramolecular forces and active-site dynamics that tune the properties of PYP's chromophore to absorb blue light, suppress fluorescence, and favor the required light-driven double-bond isomerization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sprenger, W.W., Hoff, W.D., Armitage, J.P. & Hellingwerf, K.J. The eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorption spectrum of the photoactive yellow protein. J. Bacteriol. 175, 3096–3104 (1993).
Jiang, Z. et al. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 285, 406–409 (1999).
Devanathan, S. et al. Femtosecond spectroscopic observations of initial intermediates in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila. Biophys. J. 77, 1017–1023 (1999).
Imamoto, Y., Mihara, K., Tokunaga, F. & Kataoka, M. Spectroscopic characterization of the photocycle intermediates of photoactive yellow protein. Biochemistry 40, 14336–14343 (2001).
Borgstahl, G.E., Williams, D.R. & Getzoff, E.D. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry 34, 6278–6287 (1995).
Genick, U.K. et al. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science 275, 1471–1475 (1997).
Genick, U.K., Soltis, S.M., Kuhn, P., Canestrelli, I.L. & Getzoff, E.D. Structure at 0.85 Å resolution of an early protein photocycle intermediate. Nature 392, 206–209 (1998).
Craven, C.J. et al. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in solution by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry 39, 14392–14399 (2000).
Ren, Z. et al. A molecular movie at 1.8 Å resolution displays the photocycle of photoactive yellow protein, a eubacterial blue-light receptor from nanoseconds to seconds. Biochemistry 40, 13788–13801 (2001).
Unno, M., Kumauchi, M., Sasaki, J., Tokunaga, F. & Yamauchi, S. Resonance Raman spectroscopy and quantum chemical calculations reveal structural changes in the active site of photoactive yellow protein. Biochemistry 41, 5668–5674 (2002).
Thompson, M.J., Bashford, D., Noodleman, L. & Getzoff, E.D. Photoisomerization and proton transfer in photoactive yellow protein. J. Am. Chem. Soc. 125, 8186–8194 (2003).
Baca, M. et al. Complete chemical structure of photoactive yellow protein: novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry 33, 14369–14377 (1994).
van Brederode, M.E., Gensch, T., Hoff, W.D., Hellingwerf, K.J. & Braslavsky, S.E. Photoinduced volume change and energy storage associated with the early transformations of the photoactive yellow protein from Ectothiorhodospira halophila. Biophys. J. 68 1101–1109 (1995).
Meyer, T.E., Yakali, E., Cusanovich, M.A. & Tollin, G. Properties of a water-soluble, yellow protein isolated from a halophilic phototrophic bacterium that has photochemical activity analogous to sensory rhodopsin. Biochemistry 26, 418–423 (1987).
Kim, M., Mathies, R.A., Hoff, W.D. & Hellingwerf, K.J. Resonance Raman evidence that the thioester-linked 4-hydroxycinnamyl chromophore of photoactive yellow protein is deprotonated. Biochemistry 34, 12669–12672 (1995).
Brudler, R. et al. Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein. Biochemistry 39, 13478–13486 (2000).
Demchuk, E., Genick, U.K., Woo, T.T., Getzoff, E.D. & Bashford, D. Protonation states and pH titration in the photocycle of photoactive yellow protein. Biochemistry 39, 1100–1113 (2000).
Word, J.M. et al. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol. 285, 1711–1733 (1999).
Ben-Nun, M. & Martinez, T.J. Ab initio molecular dynamics study of cis-trans photo-isomerization in ethylene. Chem. Phys. Lett. 298, 57–65 (1998).
Kobayashi, T., Saito, T. & Ohtani, H. Real-time spectroscopy of transition states in bacteriorhodopsin during retinal isomerization. Nature 414, 531–534 (2001).
Takeshita, K., Imamoto, Y., Kataoka, M., Tokunaga, F. & Terazima, M. Themodynamic and transport properties of intermediate states of the photocyclic reaction of photoactive yellow protein. Biochemistry 41, 3037–3048 (2002).
Alber, T., Gilber, W.A., Ponzi, D.R. & Petzko, G.A. The role of mobility in the substrate binding and catalytic machinery of enzymes. Ciba Found. Symp. 93, 4–24 (1983).
Agarwal, P.K., Billeter, S.R., Rajagopalan, P.T., Benkovic, S.J. & Hammes-Schiffer, S. Network of coupled promoting motions in enzyme catalysis. Proc. Natl. Acad. Sci. USA 99, 2794–2799 (2002).
Caratzoulas, S., Mincer, J.S. & Schwartz, S.D. Identification of a protein-promoting vibration in the reaction catalyzed by horse liver alcohol dehydrogenase. J. Am. Chem. Soc. 124, 3270–3276 (2002).
McRee, D.E. XtalView/XFIT—a versatile program for manipulation of atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999).
Sheldrick, G.M. & Schneider, T.R. SHELXL: high resolution refinement. Methods Enzymol. 277, 319–343 (1997).
Press, W., Teukolsky, S.A., Vetterling, W.T. & Flannery, B.P. Numerical Recipes in C: The Art of Scientific Computing edn. 2 (Cambridge Univ. Press, Cambridge, UK, 1995).
Merritt, E.A. & Bacon, D.J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Acknowledgements
We thank C. Putnam for generating the photographs in Figure 2 and 1. Canestrelli for growing the crystals used in this study. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the US Department of Energy (DOE), Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the DOE, Office of Biological and Environmental Research, and by the US National Institutes of Health (NIH), US National Center for Research Resources, Biomedical Technology Program and the US National Institute of General Medical Sciences. This work was supported through a vision research grant from the Karl Kirchgessner Foundation (U.K.G.) and a grant from the NIH (E.D.G.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Getzoff, E., Gutwin, K. & Genick, U. Anticipatory active-site motions and chromophore distortion prime photoreceptor PYP for light activation. Nat Struct Mol Biol 10, 663–668 (2003). https://doi.org/10.1038/nsb958
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb958
This article is cited by
-
High-throughput, image-based screening of pooled genetic-variant libraries
Nature Methods (2017)