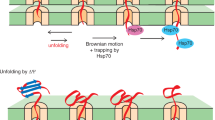
Abstract
Mitochondria can unfold importing precursor proteins by unraveling them from their N-termini. However, how this unraveling is induced is not known. Two candidates for the unfolding activity are the electrical potential across the inner mitochondrial membrane and mitochondrial Hsp70 in the matrix. Here, we propose that many precursors are unfolded by the electrical potential acting directly on positively charged amino acid side chains in the targeting sequences. Only precursor proteins with targeting sequences that are long enough to reach the matrix at the initial interaction with the import machinery are unfolded by mitochondrial Hsp70, and this unfolding occurs even in the absence of a membrane potential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Horwich, A.L., Weber-Ban, E.U. & Finley, D. Chaperone rings in protein folding and degradation. Proc. Natl. Acad. Sci. USA 96, 11033–11040 (1999).
Matouschek, A., Pfanner, N. & Voos, W. Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep. 1, 404–410 (2000).
Huang, S., Ratliff, K.S., Schwartz, M.P., Spenner, J.M. & Matouschek, A. Mitochondria unfold precursor proteins by unraveling them from their N-termini. Nature Struct. Biol. 6, 1132–1138 (1999).
Lee, C., Schwartz, M.P., Prakash, S., Iwakura, M. & Matouschek, A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell 7, 627–637 (2001).
Eilers, M. & Schatz, G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature 322, 228–232 (1986).
Schwartz, M.P., Huang, S. & Matouschek, A. The structure of precursor proteins during import into mitochondria. J. Biol. Chem. 274, 12759–12764 (1999).
Schwartz, M.P. & Matouschek, A. The dimensions of the protein import channels in the outer and inner mitochondrial membranes. Proc. Natl. Acad. Sci. USA 96, 13086–13090 (1999).
Matouschek, A. et al. Active unfolding of precursor proteins during mitochondrial protein import. EMBO J. 16, 6727–6736 (1997).
Huang, S., Murphy, S. & Matouschek, A. Effect of the protein import machinery at the mitochondrial surface on precursor stability. Proc. Natl. Acad. Sci. USA 97, 12991–12996 (2000).
Kang, P.-L. et al. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348, 137–143 (1990).
Manning-Krieg, U., Scherer, P.E. & Schatz, G. Sequential action of mitochondrial chaperones in protein import into the matrix. EMBO J. 10, 3273–3280 (1991).
Wachter, C., Schatz, G. & Glick, B.S. Protein import into mitochondria: the requirement for external ATP is precursor-specific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol. Biol. Cell 5, 465–474 (1994).
Gaume, B. et al. Unfolding of preproteins upon import into mitochondria. EMBO J. 17, 6497–6507 (1998).
Schleyer, M., Schmidt, B. & Neupert, W. Requirement of a membrane potential for the posttranslational transfer of proteins into mitochondria. Eur. J. Biochem. 125, 109–116 (1982).
Gasser, S.M., Daum, G. & Schatz, G. Import of proteins into mitochondria. Energy-dependent uptake of precursors by isolated mitochondria. J. Biol. Chem. 257, 13034–13041 (1982).
Pfanner, N. & Neupert, W. Transport of proteins into mitochondria: a potassium diffusion potential is able to drive the import of ADP/ATP carrier. EMBO J. 4, 2819–2825 (1985).
Voos, W., Gambill, B.D., Guiard, B., Pfanner, N. & Craig, E.A. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 123, 119–126 (1993).
Voos, W. et al. Differential requirement for the mitochondrial Hsp70–Tim44 complex in unfolding and translocation of preproteins. EMBO J. 15, 2668–2677 (1996).
Voisine, C. et al. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97, 565–574 (1999).
Lim, J.H., Martin, F., Guiard, B., Pfanner, N. & Voos, W. The mitochondrial Hsp70-dependent import system actively unfolds preproteins and shortens the lag phase of translocation. EMBO J. 20, 941–950 (2001).
Gambill, B.D. et al. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 123, 109–117 (1993).
Ungermann, C., Neupert, W. & Cyr, D.M. The role of hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science 266, 1250–1253 (1994).
Costanzo, M.C. et al. YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29, 75–79 (2001).
Costanzo, M.C. et al. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 28, 73–76 (2000).
Hwang, S.T., Wachter, C. & Schatz, G. Protein import into the yeast mitochondrial matrix. A new translocation intermediate between the two mitochondrial membranes. J. Biol. Chem. 266, 21083–21089 (1991).
Bauer, M.F., Sirrenberg, C., Neupert, W. & Brunner, M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87, 33–41 (1996).
Martin, J., Mahlke, K. & Pfanner, N. Role of an energized inner membrane in mitochondrial protein import. ΔΨ drives the movement of presequences. J. Biol. Chem. 266, 18051–18057 (1991).
Reers, M., Smith, T.W. & Chen, L.B. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry 30, 4480–4486 (1991).
Reers, M. et al. Mitochondrial membrane potential monitored by JC-1 dye. Methods Enzymol. 260, 406–417 (1995).
Smiley, S.T. et al. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc. Natl. Acad. Sci. USA 88, 3671–3675 (1991).
Pfanner, N. & Geissler, A. Versatility of the mitochondrial protein import machinery. Nature Rev. Mol. Cell Biol. 2, 339–349 (2001).
Brix, J., Dietmeier, K. & Pfanner, N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 272, 20730–20735 (1997).
Abe, Y. et al. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560 (2000).
Zhu, X. et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 (1996).
Ungermann, C., Guiard, B., Neupert, W. & Cyr, D.M. The ΔΨ and Hsp70/MIM44-dependent reaction cycle driving early steps of protein import into mitochondria. EMBO J. 15, 735–744 (1996).
Rudiger, S., Germeroth, L., Schneider-Mergener, J. & Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1517 (1997).
Blond-Elguindi, S. et al. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 (1993).
Chauwin, J.F., Oster, G. & Glick, B.S. Strong precursor–pore interactions constrain models for mitochondrial protein import. Biophys. J. 74, 1732–1743 (1998).
Dekker, P.J. et al. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70–Tim44. EMBO J. 16, 5408–5419 (1997).
Hartley, R.W. A two state conformational transition of the extracellular ribonuclease of Bacillus amyloliquefaciens (barnase) induced by sodium dodecyl sulfate. Biochemistry 14, 2367–2370 (1975).
Arretz, M., Schneider, H., Guiard, B., Brunner, M. & Neupert, W. Characterization of the mitochondrial processing peptidase of Neurospora crassa. J. Biol. Chem. 269, 4959–4967 (1994).
Beasley, E.M., Müller, S. & Schatz, G. The signal that sorts yeast cytochrome b 2 to the mitochondrial intermembrane space contains three distinct functional regions. EMBO J. 12, 2303–2311 (1993).
Mossakowska, D.E., Nyberg, K. & Fersht, A.R. Kinetic characterization of the recombinant ribonuclease from Bacillus amyloliquefaciens (barnase) and investigation of key residues in catalysis by site-directed mutagenesis. Biochemistry 28, 3843–3850 (1989).
Serrano, L., Kellis, J.T. Jr, Cann, P., Matouschek, A. & Fersht, A.R. The folding of an enzyme II. Substructure of barnase and the contribution of different interactions to protein stability. J. Mol. Biol. 224, 783–804 (1992).
Hartley, R.W. Directed mutagenesis and barnase–barstar recognition. Biochemistry 32, 5978–5984 (1993).
Glick, B.S. & Pon, L.A. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213–223 (1995).
Heytler, P.G. & Prichard, W.W. A new class of uncoupling agents – carbonyl cyanide phenylhydrazones. Biochem. Biophys. Res. Commun. 7, 272–275 (1962).
Geissler, A. et al. Membrane potential-driven protein import into mitochondria: the sorting sequence of cytochrome b 2 modulates the ΔΨ-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell 11, 3977–3991 (2000).
Acknowledgements
We thank C. Voisine and E. Craig, University of Wisconsin, for generously sending us the ssc1-2 and congenic wild type yeast strains; acknowledge the use of the Keck Biophysics Facility at Northwestern University; and are grateful to L. Feferman for his help with experiments. This work was funded by the National Institutes of Health. S.H. gratefully acknowledges the support by a Gramm Travel Fellowship Award from the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Huang, S., Ratliff, K. & Matouschek, A. Protein unfolding by the mitochondrial membrane potential. Nat Struct Mol Biol 9, 301–307 (2002). https://doi.org/10.1038/nsb772
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb772
This article is cited by
-
Free-energy landscapes of membrane co-translocational protein unfolding
Communications Biology (2020)
-
Role of the membrane potential in mitochondrial protein unfolding and import
Scientific Reports (2019)
-
Multistep protein unfolding during nanopore translocation
Nature Nanotechnology (2013)
-
Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force
Nature Structural & Molecular Biology (2013)
-
An energy transduction mechanism used in bacterial flagellar type III protein export
Nature Communications (2011)