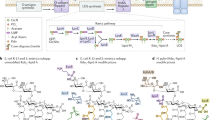
Key Points
-
The outer membrane (OM) of most Gram-negative bacteria contains lipopolysaccharide (LPS), a large molecule that contains several fatty acyl chains and up to hundreds of sugars, in its outer leaflet, which creates a barrier that prevents the entry of large polar molecules and small hydrophobic molecules.
-
The transport of millions of LPS molecules from the inner membrane (IM), across the aqueous periplasmic compartment, and across the OM to the cell surface was not well understood, except that the process is mediated by seven essential and conserved LPS transport (Lpt) proteins.
-
The extraction of LPS from the IM is mediated by an ATP binding cassette (ABC) transporter, LptB2FG, and an associated membrane protein, LptC. These proteins couple ATP hydrolysis in the cytoplasm by LptB to movement to LptC; the LptB2FG and LptB2FGC protein complexes have been purified and demonstrate ATPase activity in vitro.
-
LPS is thought to transit the periplasm by a bridge between LptC and the OM mediated by the periplasmic protein LptA. The bridge is formed by structurally homologous domains of LptC, LptA and the OM protein LptD, and it helps to mediate the transit of the hydrophobic acyl chains of LPS through an aqueous compartment.
-
The OM β-barrel protein LptD and the OM lipoprotein LptE form a two-protein plug-and-barrel complex that is responsible for transporting LPS from the periplasmic bridge across the OM to the cell surface. A current model proposes that the OM translocon changes its conformation, enabling LPS molecules to enter the barrel of LptD and move to the cell surface through lateral openings without ever residing in the inner leaflet of the OM.
-
LptD is a large β-barrel protein that contains two non-consecutive disulfide bonds, either of which is sufficient for the function of LptD. Correct rearrangement of the disulfide bonds to the final configuration is required for LptA to interact with LptD, which prevents mislocalization of LPS when the OM translocon is not properly assembled.
-
Identification of LPS transport intermediates in Escherichia coli cells has enabled the development of a system to study the ATP requirement for LPS transport out of membrane vesicles to soluble LptA. Using this system, the PEZ model was developed to describe how ATP hydrolysis by LptB in the cytoplasm 'pushes' LPS molecules in a continuous stream out of the IM toward the cell surface through the periplasmic bridge comprised of LptC, LptA and LptD.
Abstract
Gram-negative bacteria have a double-membrane cellular envelope that enables them to colonize harsh environments and prevents the entry of many clinically available antibiotics. A main component of most outer membranes is lipopolysaccharide (LPS), a glycolipid containing several fatty acyl chains and up to hundreds of sugars that is synthesized in the cytoplasm. In the past two decades, the proteins that are responsible for transporting LPS across the cellular envelope and assembling it at the cell surface in Escherichia coli have been identified, but it remains unclear how they function. In this Review, we discuss recent advances in this area and present a model that explains how energy from the cytoplasm is used to power LPS transport across the cellular envelope to the cell surface.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003).
Berg, H. C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 (2003).
Nan, B., McBride, M. J., Chen, J., Zusman, D. R. & Oster, G. Bacteria that glide with helical tracks. Curr. Biol. 24, R169–R173 (2014).
Lai, Y., Rosenshine, I., Leong, J. M. & Frankel, G. Intimate host attachment: enteropathogenic and enterohaemorrhagic Escherichia coli. Cell. Microbiol. 15, 1796–1808 (2013).
Laverty, G., Gorman, S. P. & Gilmore, B. F. Biomolecular mechanisms of Pseudomonas aeruginosa and Escherichia coli biofilm formation. Pathogens 3, 596–632 (2014).
Raetz, C. R. & Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002).
Garten, W., Hindennach, I. & Henning, U. The major proteins of the Escherichia coli outer cell-envelope membrane. Cyanogen bromide fragments of protein I, composition and order. Eur. J. Biochem. 60, 303–307 (1975).
Kamio, Y. & Nikaido, H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570 (1976). This study is the first to demonstrate that phospholipids are not exposed on the cell surface of Gram-negative bacteria.
Osborn, M. J., Gander, J. E., Parisi, E. & Carson, J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247, 3962–3972 (1972). This study demonstrates that LPS fractionates almost exclusively with the OM following sucrose-density gradient centrifugation.
Sutcliffe, I. C. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18, 464–470 (2010).
Ruiz, N., Kahne, D. & Silhavy, T. J. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 7, 677–683 (2009).
Raetz, C. R., Reynolds, C. M., Trent, M. S. & Bishop, R. E. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 (2007).
Whitfield, C. & Trent, M. S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014).
Osborn, M. J., Gander, J. E. & Parisi, E. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. J. Biol. Chem. 247, 3973–3986 (1972). This paper establishes that LPS is synthesized inside the cell, raising the question of how LPS is transported to its final destination.
Karow, M. & Georgopoulos, C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 7, 69–79 (1993).
Polissi, A. & Georgopoulos, C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 20, 1221–1233 (1996).
Zhou, Z., White, K. A., Polissi, A., Georgopoulos, C. & Raetz, C. R. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273, 12466–12475 (1998).
Doerrler, W. T., Gibbons, H. S. & Raetz, C. R. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279, 45102–45109 (2004).
Ward, A., Reyes, C. L., Yu, J., Roth, C. B. & Chang, G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl Acad. Sci. USA 104, 19005–19010 (2007).
McGrath, B. C. & Osborn, M. J. Localization of terminal steps of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173, 649–654 (1991).
Whitfield, C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 (2006).
Cuthbertson, L., Kos, V. & Whitfield, C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74, 341–362 (2010).
Greenfield, L. K. & Whitfield, C. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24 (2012).
Sperandeo, P., Deho, G. & Polissi, A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 1791, 594–602 (2009).
Simpson, B. W., May, J. M., Sherman, D. J., Kahne, D. & Ruiz, N. Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150029 (2015).
May, J. M., Sherman, D. J., Simpson, B. W., Ruiz, N. & Kahne, D. Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150027 (2015).
Sperandeo, P. et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190, 4460–4469 (2008).
Ruiz, N., Gronenberg, L. S., Kahne, D. & Silhavy, T. J. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 5537–5542 (2008).
Rietschel, E. T. et al. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8, 217–225 (1994).
Okuda, S., Freinkman, E. & Kahne, D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 (2012). This study describes the first observation of intermediate LPS transport states in living cells.
Sherman, D. J. et al. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc. Natl Acad. Sci. USA 111, 4982–4987 (2014). This paper provides the first biochemical and genetic evidence that LPS transport is powered by ATP hydrolysis by LptB.
Sperandeo, P. et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189, 244–253 (2007).
Narita, S. & Tokuda, H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583, 2160–2164 (2009). This study describes the purification of the LptBFGC complex at the IM.
Sherman, D. J., Okuda, S., Denny, W. A. & Kahne, D. Validation of inhibitors of an ABC transporter required to transport lipopolysaccharide to the cell surface in Escherichia coli. Bioorg. Med. Chem. 21, 4846–4851 (2013).
Villa, R. et al. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108 (2013).
Suits, M. D., Sperandeo, P., Deho, G., Polissi, A. & Jia, Z. Novel structure of the conserved Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 380, 476–488 (2008).
Matsuyama, S., Tajima, T. & Tokuda, H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14, 3365–3372 (1995).
Okuda, S. & Tokuda, H. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259 (2011).
Takeda, K. et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22, 3199–3209 (2003).
Okuda, S., Watanabe, S. & Tokuda, H. A short helix in the C-terminal region of LolA is important for the specific membrane localization of lipoproteins. FEBS Lett. 582, 2247–2251 (2008).
Tefsen, B., Geurtsen, J., Beckers, F., Tommassen, J. & de Cock, H. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J. Biol. Chem. 280, 4504–4509 (2005). This study provides the first evidence that LPS transport does not involve a soluble periplasmic chaperone.
Chng, S. S., Gronenberg, L. S. & Kahne, D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567 (2010). This paper provides evidence that LPS transport involves a trans-envelope bridge.
Ishidate, K. et al. Isolation of differentiated membrane domains from Escherichia coli and Salmonella typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J. Biol. Chem. 261, 428–443 (1986).
Bos, M. P., Robert, V. & Tommassen, J. Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61, 191–214 (2007).
Freinkman, E., Okuda, S., Ruiz, N. & Kahne, D. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51, 4800–4806 (2012).
Chin, J. W., Martin, A. B., King, D. S., Wang, L. & Schultz, P. G. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl Acad. Sci. USA 99, 11020–11024 (2002).
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010).
Ryu, Y. & Schultz, P. G. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3, 263–265 (2006).
Wang, L., Brock, A., Herberich, B. & Schultz, P. G. Expanding the genetic code of Escherichia coli. Science 292, 498–500 (2001).
Tran, A. X., Dong, C. & Whitfield, C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 (2010).
Bowyer, A., Baardsnes, J., Ajamian, E., Zhang, L. & Cygler, M. Characterization of interactions between LPS transport proteins of the Lpt system. Biochem. Biophys. Res. Commun. 404, 1093–1098 (2011).
Sperandeo, P. et al. New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA–LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J. Bacteriol. 193, 1042–1053 (2011).
Bos, M. P., Tefsen, B., Geurtsen, J. & Tommassen, J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl Acad. Sci. USA 101, 9417–9422 (2004).
Braun, M. & Silhavy, T. J. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45, 1289–1302 (2002).
Wu, T. et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 103, 11754–11759 (2006).
Hagan, C. L., Silhavy, T. J. & Kahne, D. β-barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 (2011).
Ruiz, N., Falcone, B., Kahne, D. & Silhavy, T. J. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317 (2005).
Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 (2003).
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005).
Sklar, J. G. et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl Acad. Sci. USA 104, 6400–6405 (2007).
Ricci, D. P. & Silhavy, T. J. The Bam machine: a molecular cooper. Biochim. Biophys. Acta 1818, 1067–1084 (2012).
Rigel, N. W. & Silhavy, T. J. Making a β-barrel: assembly of outer membrane proteins in Gram-negative bacteria. Curr. Opin. Microbiol. 15, 189–193 (2012).
Eggert, U. S. et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 294, 361–364 (2001).
Matsuyama, S., Yokota, N. & Tokuda, H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16, 6947–6955 (1997).
Okuda, S. & Tokuda, H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl Acad. Sci. USA 106, 5877–5882 (2009).
Yakushi, T., Masuda, K., Narita, S., Matsuyama, S. & Tokuda, H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2, 212–218 (2000).
Tsukahara, J., Mukaiyama, K., Okuda, S., Narita, S. & Tokuda, H. Dissection of LolB function — lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 276, 4496–4504 (2009).
Nakada, S. et al. Structural investigation of the interaction between LolA and LolB using NMR. J. Biol. Chem. 284, 24634–24643 (2009).
Chng, S. S., Ruiz, N., Chimalakonda, G., Silhavy, T. J. & Kahne, D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl Acad. Sci. USA 107, 5363–5368 (2010).
Dong, H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 (2014).
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014). Together with reference 70, this article reports the first crystal structures of LptDE.
Freinkman, E., Chng, S. S. & Kahne, D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl Acad. Sci. USA 108, 2486–2491 (2011). This paper establishes that LptD and LptE form a plug-and-barrel structure and suggests that membrane insertion proceeds through a lateral gate in a proline-rich region of the β-barrel of LptD.
Gu, Y. et al. Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD. Structure 23, 496–504 (2015).
Li, X., Gu, Y., Dong, H., Wang, W. & Dong, C. Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane. Sci. Rep. 5, 11883 (2015).
Ahn, V. E. et al. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 23, 2931–2941 (2004). This study is the first to suggest a gating mechanism by which lipids pass into the lumen of a β-barrel through a lateral opening.
Bishop, R. E. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57, 900–912 (2005).
Khan, M. A. & Bishop, R. E. Molecular mechanism for lateral lipid diffusion between the outer membrane external leaflet and a β-barrel hydrocarbon ruler. Biochemistry 48, 9745–9756 (2009).
Hearn, E. M., Patel, D. R., Lepore, B. W., Indic, M. & van den Berg, B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature 458, 367–370 (2009).
van den Berg, B. Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. Chembiochem 11, 1339–1343 (2010).
van den Berg, B., Black, P. N., Clemons, W. M. & Rapoport, T. A. Crystal structure of the long-chain fatty acid transporter FadL. Science 304, 1506–1509 (2004).
Malojcic, G. et al. LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc. Natl Acad. Sci. USA 111, 9467–9472 (2014).
Chng, S. S. et al. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 337, 1665–1668 (2012).
Ruiz, N., Chng, S. S., Hiniker, A., Kahne, D. & Silhavy, T. J. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl Acad. Sci. USA 107, 12245–12250 (2010).
Vertommen, D., Ruiz, N., Leverrier, P., Silhavy, T. J. & Collet, J. F. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9, 2432–2443 (2009).
Chimalakonda, G. et al. Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 108, 2492–2497 (2011).
Denoncin, K., Vertommen, D., Paek, E. & Collet, J. F. The protein–disulfide isomerase DsbC cooperates with SurA and DsbA in the assembly of the essential β-barrel protein LptD. J. Biol. Chem. 285, 29425–29433 (2010).
Narita, S., Masui, C., Suzuki, T., Dohmae, N. & Akiyama, Y. Protease homolog BepA (YfgC) promotes assembly and degradation of β-barrel membrane proteins in Escherichia coli. Proc. Natl Acad. Sci. USA 110, E3612–E3621 (2013).
Tran, A. X., Trent, M. S. & Whitfield, C. The LptA protein of Escherichia coli is a periplasmic lipid A-binding protein involved in the lipopolysaccharide export pathway. J. Biol. Chem. 283, 20342–20349 (2008).
Kaback, H. R. Bacterial membranes. Methods Enzymol. 22, 99–120 (1971).
Kaback, H. R. & Stadtman, E. R. Proline uptake by an isolated cytoplasmic membrane preparation of Escherichia coli. Proc. Natl Acad. Sci. USA 55, 920–927 (1966).
Kim, Y. J., Rajapandi, T. & Oliver, D. SecA protein is exposed to the periplasmic surface of the E. coli inner membrane in its active state. Cell 78, 845–853 (1994).
Masuda, K., Matsuyama, S. & Tokuda, H. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl Acad. Sci. USA 99, 7390–7395 (2002).
Ito, Y., Kanamaru, K., Taniguchi, N., Miyamoto, S. & Tokuda, H. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol. Microbiol. 62, 1064–1075 (2006).
Taniguchi, N. & Tokuda, H. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J. Biol. Chem. 283, 8538–8544 (2008).
Mizutani, M. et al. Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex. FEBS Lett. 587, 23–29 (2013).
Yasuda, M., Iguchi-Yokoyama, A., Matsuyama, S., Tokuda, H. & Narita, S. Membrane topology and functional importance of the periplasmic region of ABC transporter LolCDE. Biosci. Biotechnol. Biochem. 73, 2310–2316 (2009).
Bos, M. P. & Tommassen, J. The LptD chaperone LptE is not directly involved in lipopolysaccharide transport in Neisseria meningitidis. J. Biol. Chem. 286, 28688–28696 (2011).
Srinivas, N. et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 (2010).
Werneburg, M. et al. Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 13, 1767–1775 (2012).
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases (AI081059 to D.K.) and the National Institute of General Medical Sciences (GM034821 to T.J.S.; GM100951 to N.R.), under the US National Institutes of Health (NIH).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Periplasm
-
An aqueous, densely packed compartment between the Gram-negative inner membrane and outer membrane. The periplasm has a unique assortment of proteins and also contains a thin layer of peptidoglycan.
- Lipid A
-
The hydrophobic glucosamine-based phospholipid anchor of lipopolysaccharide molecules. Lipid A is also known as endotoxin.
- O antigen
-
Attached to the core oligosaccharide, this repetitive glycan is the outermost part of the lipopolysaccharide molecule and is a target of the host immune system.
- ATP-binding cassette transporter
-
(ABC transporter). A transmembrane protein complex that uses the energy derived from ATP binding and hydrolysis to transport various substrates. These proteins are members of one of the largest protein superfamilies and consist of transmembrane domains and conserved nucleotide-binding domains.
- Bitopic membrane protein
-
A type of membrane protein that contains only one transmembrane helix.
- β-barrel
-
A class of integral membrane protein comprised of β-strands that satisfy their peptide backbone hydrogen bonds by forming a cylindrical barrel structure, which exposes hydrophobic side chains to the membrane and shields hydrophilic side chains.
- Lipoprotein
-
A protein characterized by the presence of an amino-terminal lipid-modified cysteine that anchors the hydrophilic protein to the cell membrane.
- Transposon mutants
-
Mutants that are created through the random insertion of a transposon (or transposable element) into a genome. In the cited study, the transposable element encoded an arabinose-inducible promoter that could drive the expression of chromosomal genes located immediately downstream of the transposon insertion site. It was used to identify essential genes by screening for transposon mutants that required the presence of arabinose in the medium for growth.
- Synteny
-
The colocalization of genes in a genetic locus.
- Reductionist bioinformatics approach
-
An approach to bioinformatics in which a complex biological system is studied through a comparative analysis of similar simpler systems. In this case, a comparative bioinformatics search was used to decrease the number of candidate genes of interest by comparing the genome of interest with other genomes of smaller size.
- β-jellyroll
-
A structure in which anti-parallel β-strands are 'wrapped' into a cylindrical, barrel-like shape without necessarily maintaining a continuous hydrogen bonding network.
- Periplasmic chaperone
-
A periplasmic protein that prevents macromolecules from aggregating and assists them in reaching their destination.
- Spheroplasts
-
Osmotically fragile bacterial cells that have had their outer membranes and peptidoglycan layers incompletely disrupted, which causes them to form a spherical shape.
- Photo-crosslinking
-
The light-induced formation of a covalent bond between two molecules to detect molecular interactions.
- Unnatural amino acid
-
Non-coded, non-proteinogenic amino acid that, when incorporated into proteins, enables various new functions.
- Size-exclusion chromatography
-
A chromatographic technique that is used for preparative or analytical purposes to separate molecules (usually macromolecules) based on their size.
- β-barrel assembly machine pathway
-
(Bam pathway). Following secretion from the inner membrane and translocation across the periplasm, the Bam complex is responsible for folding and inserting β-barrel proteins into the outer membrane. In Escherichia coli, the Bam complex is composed of one β-barrel protein, BamA, and four outer membrane lipoproteins, BamB, BamC, BamD and BamE.
- Localization of lipoproteins pathway
-
(Lol pathway). A chaperone-based transport pathway that is involved in transporting outer membrane lipoproteins from the outer leaflet of the inner membrane to the inner leaflet of the outer membrane.
- Trypsin
-
A serine protease that hydrolyses peptide bonds on the carboxy-terminal side of lysine and arginine residues, and is commonly used to determine the stability of proteins.
- Crenellated β-barrel
-
A β-barrel protein in which the formation of inter-strand hydrogen bonds is disrupted, creating openings similar to the crenels in the turret of a castle.
- Right-side-out membrane vesicles
-
Membrane vesicles with a native orientation that are prepared by the osmotic lysis of spheroplasts.
- Vanadate
-
Sodium orthovanadate (Na3VO4) inhibits protein tyrosine phosphatases, alkaline phosphatases and many ATPases by acting as a phosphate analogue and binding in the active site in which phosphate usually binds.
- Peptidomimetic
-
Chemical compounds that mimic natural peptides because of their small protein-like chains.
Rights and permissions
About this article
Cite this article
Okuda, S., Sherman, D., Silhavy, T. et al. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14, 337–345 (2016). https://doi.org/10.1038/nrmicro.2016.25
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.25
This article is cited by
-
A new antibiotic traps lipopolysaccharide in its intermembrane transporter
Nature (2024)
-
Conformational Investigation of the Asymmetric Periplasmic Domains of E. coli LptB2FGC Using SDSL CW EPR Spectroscopy
Applied Magnetic Resonance (2024)
-
Surveying membrane landscapes: a new look at the bacterial cell surface
Nature Reviews Microbiology (2023)
-
Mechanism of outer membrane destabilization by global reduction of protein content
Nature Communications (2023)
-
Gut dysbiosis induces the development of mastitis through a reduction in host anti-inflammatory enzyme activity by endotoxemia
Microbiome (2022)