Key Points
-
Overlapping gene silencing mechanisms function to epigenetically repress transposable elements (TEs), including chromatin and DNA modification as well as RNAi.
-
TEs are a major component of constitutive heterochromatin at the centromere and telomere, and the regulation of these TEs is required for proper chromosome function.
-
TEs can function as the nucleation centers for facultative heterochromatin, as well as acting as boundaries to the spread of gene silencing.
-
Epialleles are formed due to the proximity of a gene to a TE, and the epigenetic regulation that the TE recruits.
-
TEs could have served as the building blocks for epigenetic phenomena such as X-chromosome inactivation, gene imprinting and V(D)J recombination.
-
The epigenetic control of transposable elements has had an important role in both organism and genome evolution.
-
How differing epigenetic control of TEs is established in genetically identical cells (variegation) remains a major question in understanding how TEs influence the genome.
Abstract
Overlapping epigenetic mechanisms have evolved in eukaryotic cells to silence the expression and mobility of transposable elements (TEs). Owing to their ability to recruit the silencing machinery, TEs have served as building blocks for epigenetic phenomena, both at the level of single genes and across larger chromosomal regions. Important progress has been made recently in understanding these silencing mechanisms. In addition, new insights have been gained into how this silencing has been co-opted to serve essential functions in 'host' cells, highlighting the importance of TEs in the epigenetic regulation of the genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Girard, L. & Freeling, M. Regulatory changes as a consequence of transposon insertion. Dev. Genet. 25, 291–296 (1999).
McClintock, B. Components of action of the regulators Spm and Ac. Carnegie Inst. Wash. Year Book 64, 527–536 (1965).
Griffith, J. L. et al. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164, 867–879 (2003).
Vastenhouw, N. L. et al. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 13, 1311–1316 (2003).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999).
Sijen, T. & Plasterk, R. H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426, 310–314 (2003). This report demonstrates that the endogenous RNAi system functions to repress TE activity in a tissue- and developmentally specific manner.
Watanabe, T. et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732–1743 (2006). This report confirms the previous findings in non-mammalian eukaryotes that TEs are an important source and target of siRNAs.
Yang, N. & Kazazian, H. H. Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature Struct. Mol. Biol. 13, 763–771 (2006). This publication demonstrated that siRNAs are generated from TEs in human cells, as well as showing that the LINE1 antisense promoter is essential for the production of these siRNAs.
Lippman, Z., May, B., Yordan, C., Singer, T. & Martienssen, R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1, e67 (2003).
Gendrel, A. V., Lippman, Z., Yordan, C., Colot, V. & Martienssen, R. A. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873 (2002).
Martens, J. H. et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800–812 (2005).
Walsh, C. P., Chaillet, J. R. & Bestor, T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet. 20, 116–117 (1998).
Woodcock, D. M., Lawler, C. B., Linsenmeyer, M. E., Doherty, J. P. & Warren, W. D. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J. Biol. Chem. 272, 7810–7816 (1997).
Bourc'his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004). This study provides evidence of the tissue-specific de novo methylation of retrotransposons in mammals, as well as demonstrating that this methylation is essential for viability.
Lippman, Z. et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476 (2004). This study describes the microarray analysis of an entire chromosomal knob, its TE composition and its dependency on DNA methylation and chromatin remodelling.
Kato, M., Takashima, K. & Kakutani, T. Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana. Genetics 168, 961–969 (2004).
Yan, Q., Cho, E., Lockett, S. & Muegge, K. Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol. Cell. Biol. 23, 8416–8428 (2003).
Huang, J. et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 32, 5019–5028 (2004). This work demonstrates that the SWI/SNF Lsh1 gene regulates only repetitive DNA such as TEs, as well as showing the large amount of the genome that is mis-expressed when TE silencing is lost.
Martienssen, R. A., Zaratiegui, M. & Goto, D. B. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 21, 450–456 (2005).
Buhler, M., Verdel, A. & Moazed, D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125, 873–886 (2006).
Irvine, D. V. et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science 313, 1134–1137 (2006). This report demonstrates that the spread of chromatin modifications is dependent on read-through transcription and the catalytic activity of argonaute.
Hansen, K. R. et al. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 25, 590–601 (2005).
Mochizuki, K. & Gorovsky, M. A. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 14, 181–187 (2004).
Xie, Z. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, e104 (2004).
Hamilton, A., Voinnet, O., Chappell, L. & Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 (2002). This is the first correlation between the TE-produced longer class of siRNAs in plants and their role in DNA methylation.
Qi, Y. et al. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443, 1008–1012 (2006).
Herr, A. J., Jensen, M. B., Dalmay, T. & Baulcombe, D. C. RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120 (2005).
Onodera, Y. et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622 (2005).
Huettel, B. et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 25, 2828–2836 (2006).
Pontier, D. et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19, 2030–2040 (2005).
Kanno, T. et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nature Genet. 37, 761–765 (2005).
Li, C. F. et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126, 93–106 (2006).
Pontes, O. et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126, 79–92 (2006).
Kanno, T. et al. A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO Rep. 6, 649–655 (2005).
Matzke, M. A. & Birchler, J. A. RNAi-mediated pathways in the nucleus. Nature Rev. Genet. 6, 24–35 (2005).
Cao, X. & Jacobsen, S. E. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12, 1138–1144 (2002).
Weinberg, M. S. et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12, 256–262 (2006).
Castro, J. P. & Carareto, C. M. Drosophila melanogaster P transposable elements: mechanisms of transposition and regulation. Genetica 121, 107–118 (2004).
Reiss, D., Josse, T., Anxolabehere, D. & Ronsseray, S. aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol. Genet. Genomics 272, 336–343 (2004). This report was the first to link an argonaute gene and RNA-based silencing to the trans silencing signal in TE cytotype control.
Kavi, H. H., Fernandez, H. R., Xie, W. & Birchler, J. A. RNA silencing in Drosophila. FEBS Lett. 579, 5940–5949 (2005).
Jensen, S., Gassama, M. P. & Heidmann, T. Taming of transposable elements by homology-dependent gene silencing. Nature Genet. 21, 209–212 (1999). This important report shows that a short, non-protein-encoding fragment of a TE can suppress hybrid dysgenesis and transposition activity when the full-length TE enters a naïve genome.
Vagin, V. V. et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006). This recent report shows that rasiRNAs are structurally different from siRNAs, and they associate with Piwi to silence TEs in D. melanogaster.
Sarot, E., Payen-Groschene, G., Bucheton, A. & Pelisson, A. Evidence for a Piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166, 1313–1321 (2004).
Rehwinkel, J. et al. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 26, 2965–2975 (2006).
Cox, D. N. et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (1998).
Klattenhoff, C. et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55 (2007).
Aravin, A. et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006).
Grivna, S. T., Beyret, E., Wang, Z. & Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714 (2006).
Girard, A., Sachidanandam, R., Hannon, G. J. & Carmell, M. A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006).
Aravin, A. A. et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 5, 337–350 (2003).
Motamedi, M. R. et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 (2004).
Schueler, M. G. & Sullivan, B. A. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genomics Hum. Genet. 7, 301–313 (2006).
The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Dawe, R. K. & Henikoff, S. Centromeres put epigenetics in the driver's seat. Trends Biochem. Sci. 31, 662–669 (2006).
Peters, A. H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001).
De La Fuente, R. et al. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nature Cell Biol. 8, 1448–1454 (2006).
Pidoux, A. L. & Allshire, R. C. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 569–579 (2005).
Lippman, Z. & Martienssen, R. The role of RNA interference in heterochromatic silencing. Nature 431, 364–370 (2004).
May, B. P., Lippman, Z. B., Fang, Y., Spector, D. L. & Martienssen, R. A. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 1, e79 (2005).
Jiang, J., Birchler, J. A., Parrott, W. A. & Dawe, R. K. A molecular view of plant centromeres. Trends Plant Sci. 8, 570–575 (2003).
Fukagawa, T. et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nature Cell Biol. 6, 784–791 (2004).
Kanellopoulou, C. et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 (2005). This report confirms that, as in lower eukaryotes, dicer and small-RNA-based mechanisms are responsible for centromere condensation in mammals.
Ohzeki, J., Nakano, M., Okada, T. & Masumoto, H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775 (2002).
Wong, L. H. & Choo, K. H. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 20, 611–616 (2004).
Martienssen, R. A. Maintenance of heterochromatin by RNA interference of tandem repeats. Nature Genet. 35, 213–214 (2003).
Nakamura, T. M. et al. Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959 (1997).
Pardue, M. L. et al. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 13, 443–453 (2005).
Casacuberta, E. & Pardue, M. L. RNA interference has a role in regulating Drosophila telomeres. Genome Biol. 7, 220 (2006).
Savitsky, M., Kwon, D., Georgiev, P., Kalmykova, A. & Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 20, 345–354 (2006).
Gonzalo, S. et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nature Cell Biol. 8, 416–424 (2006).
Schotta, G., Ebert, A., Dorn, R. & Reuter, G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14, 67–75 (2003).
Sun, F. L. et al. cis-acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 24, 8210–8220 (2004).
Haynes, K. A., Caudy, A. A., Collins, L. & Elgin, S. C. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr. Biol. 16, 2222–2227 (2006).
Schotta, G., Ebert, A. & Reuter, G. SU(VAR)3–9 is a conserved key function in heterochromatic gene silencing. Genetica 117, 149–158 (2003).
Gaszner, M. & Felsenfeld, G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nature Rev. Genet. 7, 703–713 (2006).
Lei, E. P. & Corces, V. G. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nature Genet. 38, 936–941 (2006).
Lyon, M. F. Do LINEs have a role in X-chromosome inactivation? J. Biomed. Biotechnol. 2006, 59746 (2006).
Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005).
Cohen, D. E. et al. The DXPas34 repeat regulates random and imprinted X inactivation. Dev. Cell 12, 57–71 (2007).
Rakyan, V. K., Blewitt, M. E., Druker, R., Preis, J. I. & Whitelaw, E. Metastable epialleles in mammals. Trends Genet. 18, 348–351 (2002).
Jordan, I. K., Rogozin, I. B., Glazko, G. V. & Koonin, E. V. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72 (2003).
Romanish, M. T., Lock, W. M., de Lagemaat, L. N., Dunn, C. A. & Mager, D. L. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 3, e10 (2007).
Garfinkel, D. J. Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet. Genome Res. 110, 63–69 (2005).
Nigumann, P., Redik, K., Matlik, K. & Speek, M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79, 628–634 (2002). This paper reports the discovery that tissue-specific transcripts that are generated from the outward-reading antisense promoter of LINE1 express neighbouring genes in large numbers.
Hodgetts, R. B. & O'Keefe, S. L. The mutant phenotype associated with P-element alleles of the vestigial locus in Drosophila melanogaster may be caused by a readthrough transcript initiated at the P-element promoter. Genetics 157, 1665–1672 (2001).
Morgan, H. D., Sutherland, H. G., Martin, D. I. & Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genet. 23, 314–318 (1999). This landmark report demonstrates that a mosaic coat-colour pattern is defined by the epigenetic state of a nearby TE, and that these epigenetic patterns are not fully erased when passed through meiosis, creating heritable patterns of epigenetic gene expression.
Blewitt, M. E., Vickaryous, N. K., Paldi, A., Koseki, H. & Whitelaw, E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2, e49 (2006).
Gaudet, F. et al. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol. Cell. Biol. 24, 1640–1648 (2004).
Matlik, K., Redik, K. & Speek, M. L1 antisense promoter drives tissue-specific transcription of human genes. J. Biomed. Biotechnol. 2006, 71753 (2006).
Peaston, A. E. et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606 (2004).
Martienssen, R. & Baron, A. Coordinate suppression of mutations caused by Robertson's mutator transposons in maize. Genetics 136, 1157–1170 (1994).
Whitelaw, E. & Martin, D. I. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nature Genet. 27, 361–365 (2001). In this provocative commentary, it is suggested that the differential metastable epigenetic silencing of TEs that are scattered throughout the genome produce intraspecies diversity.
Walker, E. L. Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148, 1973–1981 (1998).
Stam, M., Belele, C., Dorweiler, J. E. & Chandler, V. L. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16, 1906–1918 (2002).
Alleman, M. et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442, 295–298 (2006).
Woodhouse, M. R., Freeling, M. & Lisch, D. Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4, e339 (2006).
McGinnis, K. M., Springer, C., Lin, Y., Carey, C. C. & Chandler, V. Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics 173, 1637–1647 (2006).
Rassoulzadegan, M. et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474 (2006).
Rubin, C. M., VandeVoort, C. A., Teplitz, R. L. & Schmid, C. W. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. 22, 5121–5127 (1994).
Allen, E. et al. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl Acad. Sci. USA 100, 9940–9945 (2003). In this unique survey of the genome, LINE elements were found to be significantly associated with imprinted genes.
Greally, J. M. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl Acad. Sci. USA 99, 327–332 (2002).
Reik, W. & Lewis, A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nature Rev. Genet. 6, 403–410 (2005).
Bourc'his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Youngson, N. A., Kocialkowski, S., Peel, N. & Ferguson-Smith, A. C. A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61, 481–490 (2005).
Ono, R. et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237 (2001).
Davis, E. et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15, 743–749 (2005).
Chan, S. W., Zhang, X., Bernatavichute, Y. V. & Jacobsen, S. E. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 4, e363 (2006).
Kinoshita, T. et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303, 521–523 (2004).
Walter, J., Hutter, B., Khare, T. & Paulsen, M. Repetitive elements in imprinted genes. Cytogenet. Genome Res. 113, 109–115 (2006).
Sen, R. & Oltz, E. Genetic and epigenetic regulation of IgH gene assembly. Curr. Opin. Immunol. 18, 237–242 (2006).
Bolland, D. J. et al. Antisense intergenic transcription in V(D)J recombination. Nature Immunol. 5, 630–637 (2004).
Kidwell, M. G. & Lisch, D. R. Transposable elements and host genome evolution. Trends Ecol. Evol. 15, 95–99 (2000).
Hickey, D. A. Selfish DNA: a sexually-transmitted nuclear parasite. Genetics 101, 519–531 (1982).
Doolittle, W. F. & Sapienza, C. Selfish genes, the phenotype paradigm and genome evolution. Nature 284, 601–603 (1980).
Wang, Q. & Dooner, H. K. Eukaryotic transposable elements and genome evolution special feature: remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl Acad. Sci. USA 103, 17644–17649 (2006).
Boissinot, S., Davis, J., Entezam, A., Petrov, D. & Furano, A. V. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl Acad. Sci. USA 103, 9590–9594 (2006).
Seleme Mdel, C. et al. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc. Natl Acad. Sci. USA 103, 6611–6616 (2006). This report demonstrates that there is a surprisingly high level of individual variation and TE-activity potential between humans.
Deininger, P. L., Moran, J. V., Batzer, M. A. & Kazazian, H. H. Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 13, 651–658 (2003).
Sandovici, I. et al. Interindividual variability and parent of origin DNA methylation differences at specific human Alu elements. Hum. Mol. Genet. 14, 2135–2143 (2005).
Rangwala, S. H. et al. Meiotically stable natural epialleles of Sadhu, a novel Arabidopsis retroposon. PLoS Genet. 2, e36 (2006).
Kazazian, H. H. Jr. Mobile elements: drivers of genome evolution. Science 303, 1626–1632 (2004).
Slotkin, R. K., Freeling, M. & Lisch, D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nature Genet. 37, 641–644 (2005).
Ronsseray, S., Josse, T., Boivin, A. & Anxolabehere, D. Telomeric transgenes and trans-silencing in Drosophila. Genetica 117, 327–335 (2003).
Axtell, M. J., Jan, C., Rajagopalan, R. & Bartel, D. P. A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577 (2006).
Schlappi, M., Raina, R. & Fedoroff, N. Epigenetic regulation of the maize Spm transposable element: novel activation of a methylated promoter by TnpA. Cell 77, 427–437 (1994).
McClintock, B. The significance of responses of the genome to challenge. Science 226, 792–801 (1984).
Capy, P., Gasperi, G., Biemont, C. & Bazin, C. Stress and transposable elements: co-evolution or useful parasites? Heredity 85, 101–106 (2000).
Allshire, R. C., Javerzat, J. P., Redhead, N. J. & Cranston, G. Position effect variegation at fission yeast centromeres. Cell 76, 157–169 (1994).
Kidwell, M. G. Reciprocal differences in female recombination associated with hybrid dysgenesis in Drosophila melanogaster. Genet. Res. 30, 77–88 (1977).
Vieira, C., Aubry, P., Lepetit, D. & Biemont, C. A temperature cline in copy number for 412 but not roo/B104 retrotransposons in populations of Drosophila simulans. Proc. Biol. Sci. 265, 1161–1165 (1998).
Jia, S., Noma, K. & Grewal, S. I. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971–1976 (2004).
Wolff, G. L., Kodell, R. L., Moore, S. R. & Cooney, C. A. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 12, 949–957 (1998).
Cooney, C. A., Dave, A. A. & Wolff, G. L. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 132, 2393S–2400S (2002). This study demonstrates the surprising result that dietary supplementation of a methylation donor molecule heritably alters a TE-induced epiallele that confers coat colour in mice.
Waterland, R. A. & Jirtle, R. L. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300 (2003).
Ivics, Z., Hackett, P. B., Plasterk, R. H. & Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 (1997).
Miskey, C., Izsvak, Z., Plasterk, R. H. & Ivics, Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31, 6873–6881 (2003).
An, W. et al. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl Acad. Sci. USA 103, 18662–18667 (2006).
Biemont, C. & Vieira, C. What transposable elements tell us about genome organization and evolution: the case of Drosophila. Cytogenet. Genome Res. 110, 25–34 (2005).
Kidwell, M. G. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115, 49–63 (2002).
Hawkins, J. S., Kim, H., Nason, J. D., Wing, R. A. & Wendel, J. F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 16, 1252–1261 (2006).
Piegu, B. et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16, 1262–1269 (2006).
Iida, S., Morita, Y., Choi, J. D., Park, K. I. & Hoshino, A. Genetics and epigenetics in flower pigmentation associated with transposable elements in morning glories. Adv. Biophys. 38, 141–159 (2004).
Martienssen, R. A. & Colot, V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293, 1070–1074 (2001).
Kapitonov, V. V. & Jurka, J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3, e181 (2005). This study provides the missing evidence of the TE-family origin of the RAG transposase proteins.
Acknowledgements
The authors apologize if relevant publications were not cited due to space constraints. R.K.S is supported by a postdoctoral fellowship from the US National Institutes of Health. Work in the authors' laboratory is supported by a grant from the National Institutes of Health to R.M.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
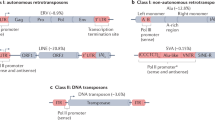
- Transposable elements
-
All mobile DNA segments in the genome, regardless of their mechanism of transposition.
- Transpose
-
The movement of a genetic element from one location of the genome to another.
- Autonomous element
-
A transposable element that produces all the proteins that are required for transposition.
- Cryptic element
-
A transposable element that is epigenetically inactivated for a period of time and has lost the ability to mobilize.
- Epigenetic
-
A heritable change that is not caused by a genetic mutation.
- Non-autonomous element
-
A transposable element that does not produce the proteins required for transposition. These elements are dependent on the proteins produced by autonomous elements of the same element family to transpose.
- Heterochromatin
-
The portion of interphase chromosomes that remains densely stained and condensed after cell division. Heterochromatic regions are generally gene poor, replicate late, and have a low level of transcription.
- Imprinting
-
A gene is expressed from only one allele in a diploid organism, depending on the parent of origin.
- Duplicative transposition
-
A mechanism of transposition that results in a copy of the element at both the excision and acceptor site. This can occur even after excision of the element by the process of gap repair.
- Cut-and-paste transposition
-
A mechanism of transposition in which no copy of the element remains at the excision site.
- Gap repair
-
A process whereby, after excision of a DNA transposon, the element can be copied back into the excision site from the template of the sister chromatid or homologous chromosome.
- DNA transposon
-
Transposable elements that do not use a reverse-transcription step to integrate copies into the genome.
- Terminal inverted repeat
-
Repeats that flank most DNA transposons and lie in an inverted orientation.
- Retrotransposon
-
Transposable elements that use a reverse-transcription step to integrate copies into the genome. Also known as retroposons.
- Presetting
-
Describes the inheritance of transposable-element
- Position effect variegation
-
Unstable gene-expression patterns that are determined by the location of a gene.
- LTR retrotransposon
-
Long terminal repeat retrotransposons are a type of retrotransposon that has repeats in a direct orientation at either end, in contrast with non-LTR retrotransposons.
- Transposable element cycling
-
The transformation of a transposable element from an epigenetically inactive state to an active state, and back to an epigenetically inactive state, over the course of development or over multiple generations.
- Polycomb group
-
A class of proteins, originally described in Drosophila melanogaster, the function of which is to maintain stable and heritable epigenetic repression.
- non-LTR retrotransposon
-
A retrotransposon that is not flanked by long terminal repeats.
- Constitutive heterochromatin
-
Heterochromatin that is heritable and is found at a particular locus at all times.
- Facultative heterochromatin
-
Heterochromatin that switches to euchromatin at points during development.
Rights and permissions
About this article
Cite this article
Slotkin, R., Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8, 272–285 (2007). https://doi.org/10.1038/nrg2072
Issue Date:
DOI: https://doi.org/10.1038/nrg2072
This article is cited by
-
The human PTGR1 gene expression is controlled by TE-derived Z-DNA forming sequence cooperating with miR-6867-5p
Scientific Reports (2024)
-
Molecular mechanisms and regulation of recombination frequency and distribution in plants
Theoretical and Applied Genetics (2024)
-
Investigation of Tos17 LTR retrotransposon movements in rice (Oryza sativa L.) under nickel and boron stress
Cereal Research Communications (2024)
-
Emerging roles of tRNA-derived fragments in cancer
Molecular Cancer (2023)
-
Lineage-specific amplification and epigenetic regulation of LTR-retrotransposons contribute to the structure, evolution, and function of Fabaceae species
BMC Genomics (2023)