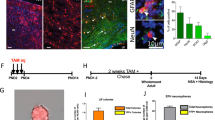
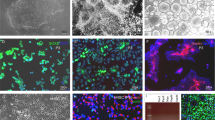
Abstract
Endogenous neural stem cells (NSCs) in the adult hippocampus are considered to be bi-potent, as they only produce neurons and astrocytes in vivo. In mouse, we found that inactivation of neurofibromin 1 (Nf1), a gene mutated in neurofibromatosis type 1, unlocked a latent oligodendrocyte lineage potential to produce all three lineages from NSCs in vivo. Our results suggest an avenue for promoting stem cell plasticity by targeting barriers of latent lineage potential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gage, F.H. Science 287, 1433–1438 (2000).
Ming, G.L. & Song, H. Neuron 70, 687–702 (2011).
Menn, B. et al. J. Neurosci. 26, 7907–7918 (2006).
Calzolari, F. et al. Nat. Neurosci. 18, 490–492 (2015).
Ortega, F. et al. Nat. Cell Biol. 15, 602–613 (2013).
Gutmann, D.H., Parada, L.F., Silva, A.J. & Ratner, N. J. Neurosci. 32, 14087–14093 (2012).
Li, Y., Li, Y., McKay, R.M., Riethmacher, D. & Parada, L.F. J. Neurosci. 32, 3529–3539 (2012).
Lagace, D.C. et al. J. Neurosci. 27, 12623–12629 (2007).
Dranovsky, A. et al. Neuron 70, 908–923 (2011).
Bonaguidi, M.A. et al. Cell 145, 1142–1155 (2011).
Song, J. et al. Nature 489, 150–154 (2012).
Jang, M.H. et al. Cell Stem Cell 12, 215–223 (2013).
Wang, Y. et al. Cell 150, 816–830 (2012).
Liu, C. et al. Cell 146, 209–221 (2011).
Chetty, S. et al. Mol. Psychiatry 19, 1275–1283 (2014).
Ahn, S. & Joyner, A.L. Nature 437, 894–897 (2005).
Kriegstein, A. & Alvarez-Buylla, A. Annu. Rev. Neurosci. 32, 149–184 (2009).
Jessberger, S., Toni, N., Clemenson, G.D. Jr., Ray, J. & Gage, F.H. Nat. Neurosci. 11, 888–893 (2008).
Magnusson, J.P. et al. Science 346, 237–241 (2014).
Nato, G. et al. Development 142, 840–845 (2015).
Balordi, F. & Fishell, G. J. Neurosci. 27, 14248–14259 (2007).
Novak, A., Guo, C., Yang, W., Nagy, A. & Lobe, C.G. Genesis 28, 147–155 (2000).
Zhu, Y. et al. Genes Dev. 15, 859–876 (2001).
Berg, D.A. et al. Front. Biol. 10, 262–271 (2015).
Sun, G.J. et al. Proc. Natl. Acad. Sci. USA 112, 9484–9489 (2015).
Acknowledgements
We thank members of the Ming and Song laboratories for discussion, and Y. Cai and L. Liu for technical support. This work was supported by the US National Institutes of Health (NS047344 to H.S., NS080913 to M.A.B., and NS048271 and MH105128 to G.M.), and by a pre-doctoral fellowship from The Children's Tumor Foundation to G.J.S.
Author information
Authors and Affiliations
Contributions
G.J.S., H.S. and G.M. designed the project. G.J.S. contributed to all aspects of the study. Y. Zhou, S.I., G.S.-O'B., N.K.K. and N.M. performed the experiments. M.A.B. provided some initial data. Y. Zhu contributed reagents. G.J.S., H.S. and G.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Inducible recombination and cell targeting of Nf1 inactivation in NSCs at the population level using Nestin-CreERT2 mice.
(a) Genetic recombination model for conditional inactivation of Nf1 in the adult mouse brain. (b) Injection timeline and paradigm for population analysis using NestinCreERT2-based cell targeting. (c) Sample confocal image of GFP+ cells in Nf1Nestin animal at 1 mpi. Arrowheads point to GFP+MCM2+NG2+ OPCs. (d) Sample confocal image of an ectopically Olig2-expressing RGL in Nf1Nestin animal at 14 dpi. Scale bars: 10 μm.
Supplementary Figure 2 Inducible recombination and cell targeting of Nf1 inactivation in NSCs at the clonal level using Nestin-CreERT2 mice.
(a) Injection timeline and paradigm for clonal analysis using NestinCreERT2-based cell targeting. (b) Cumulative distribution plot of cell number of GFP+ clones for those containing OPCs in Nf1Nestin animals (solid circle) and those in controlNestin animals (open circle) at 1 mpi. Each symbol represents data from one clone. *p < 0.01, two-sample Kolmogorov–Smirnov test. (c) Clone at 1 mpi in Nf1Nestin animal with Prox1+ newborn dentate granule neurons (N1, N2) ectopically migrated into the molecular layer (ML) of the dentate gyrus. ML, molecular layer; GCL, granule cell layer; SGZ, subgranular zone. Scale bar: 10 μm.
Supplementary Figure 3 Clonal lineage tracing of RGLs upon conditional Nf1 inactivation using Gli1CreERT2-based cell targeting.
(a) Injection timeline and paradigm for clonal analysis using Gli1CreERT2-based cell targeting for adult mice. (b) A tri-lineage clone at 1 mpi in Nf1Gli1 animal with lost or differentiated RGL, astrocytes (open arrowheads), OPCs (closed arrowheads), and mature newborn neurons. Right: high magnification confocal image of boxed region denoting NG2+ OPC. Scale bars: 50 μm and 10 μm (right). (c) A clone at 1 mpi in controlGli1 animal with a maintained RGL (boxed) and mature newborn neurons. Right: maintained nestin+ RGL. Scale bar: 10 μm. See Supplementary Table 3 for number of hemispheres examined under different conditions.
Supplementary Figure 4 Models of adult NSC fate regulation.
Under normal conditions, endogenous RGLs in the adult mouse dentate gyrus are bi-potent and generate only neurons and astrocytes in vivo. NF1 suppresses the oligodendrocytic lineage potential, which can be revealed by removal of NF1.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 1–4 (PDF 953 kb)
3D rendering and fly-through of reconstructed 2 mpi Nf1Nestin clone shown in Figure 2a.
Four representative OPCs are labeled in addition to the mother RGL. OPC, oligodendrocyte progenitor cell; RGL, radial glia-like cell (MOV 11862 kb)
Rights and permissions
About this article
Cite this article
Sun, G., Zhou, Y., Ito, S. et al. Latent tri-lineage potential of adult hippocampal neural stem cells revealed by Nf1 inactivation. Nat Neurosci 18, 1722–1724 (2015). https://doi.org/10.1038/nn.4159
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4159