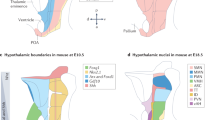
Abstract
The hypothalamus is a central regulator of many behaviors that are essential for survival, such as temperature regulation, food intake and circadian rhythms. However, the molecular pathways that mediate hypothalamic development are largely unknown. To identify genes expressed in developing mouse hypothalamus, we performed microarray analysis at 12 different developmental time points. We then conducted developmental in situ hybridization for 1,045 genes that were dynamically expressed over the course of hypothalamic neurogenesis. We identified markers that stably labeled each major hypothalamic nucleus over the entire course of neurogenesis and constructed a detailed molecular atlas of the developing hypothalamus. As a proof of concept of the utility of these data, we used these markers to analyze the phenotype of mice in which Sonic Hedgehog (Shh) was selectively deleted from hypothalamic neuroepithelium and found that Shh is essential for anterior hypothalamic patterning. Our results serve as a resource for functional investigations of hypothalamic development, connectivity, physiology and dysfunction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
O'Rahilly, S. Human genetics illuminates the paths to metabolic disease. Nature 462, 307–314 (2009).
Caqueret, A., Yang, C., Duplan, S., Boucher, F. & Michaud, J.L. Looking for trouble: a search for developmental defects of the hypothalamus. Horm. Res. 64, 222–230 (2005).
Fyffe, S.L. et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression and the response to stress. Neuron 59, 947–958 (2008).
McGill, B.E. et al. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA 103, 18267–18272 (2006).
Lee, J.E., Wu, S.F., Goering, L.M. & Dorsky, R.I. Canonical Wnt signaling through Lef1 is required for hypothalamic neurogenesis. Development 133, 4451–4461 (2006).
Dale, J.K. et al. Cooperation of BMP7 and SHH in the induction of forebrain ventral midline cells by prechordal mesoderm. Cell 90, 257–269 (1997).
Manning, L. et al. Regional morphogenesis in the hypothalamus: a BMP-Tbx2 pathway coordinates fate and proliferation through Shh downregulation. Dev. Cell 11, 873–885 (2006).
Ohyama, K., Das, R. & Placzek, M. Temporal progression of hypothalamic patterning by a dual action of BMP. Development 135, 3325–3331 (2008).
Shimada, M. & Nakamura, T. Time of neuron origin in mouse hypothalamic nuclei. Exp. Neurol. 41, 163–173 (1973).
Michaud, J.L., Rosenquist, T., May, N.R. & Fan, C.M. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 12, 3264–3275 (1998).
Schonemann, M.D. et al. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9, 3122–3135 (1995).
Labosky, P.A. et al. The winged helix gene, Mf3, is required for normal development of the diencephalon and midbrain, postnatal growth and the milk-ejection reflex. Development 124, 1263–1274 (1997).
Wehr, R., Mansouri, A., de Maeyer, T. & Gruss, P. Fkh5-deficient mice show dysgenesis in the caudal midbrain and hypothalamic mammillary body. Development 124, 4447–4456 (1997).
Davis, A.M. et al. Loss of steroidogenic factor 1 alters cellular topography in the mouse ventromedial nucleus of the hypothalamus. J. Neurobiol. 60, 424–436 (2004).
Acampora, D. et al. Progressive impairment of developing neuroendocrine cell lineages in the hypothalamus of mice lacking the Orthopedia gene. Genes Dev. 13, 2787–2800 (1999).
Goshu, E. et al. Sim2 contributes to neuroendocrine hormone gene expression in the anterior hypothalamus. Mol. Endocrinol. 18, 1251–1262 (2004).
Gray, P.A. et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306, 2255–2257 (2004).
Alvarez-Bolado, G. & Eichele, G. Analysing the developing brain transcriptome with the GenePaint platform. J. Physiol. (Lond.) 575, 347–352 (2006).
Elmquist, J.K., Elias, C.F. & Saper, C.B. From lesions to leptin: hypothalamic control of food intake and body weight. Neuron 22, 221–232 (1999).
Charlton, H. Hypothalamic control of anterior pituitary function: a history. J. Neuroendocrinol. 20, 641–646 (2008).
Kruk, M.R. et al. The hypothalamus: cross-roads of endocrine and behavioural regulation in grooming and aggression. Neurosci. Biobehav. Rev. 23, 163–177 (1998).
Saper, C.B., Scammell, T.E. & Lu, J. Hypothalamic regulation of sleep and circadian rhythms. Nature 437, 1257–1263 (2005).
Ulrich-Lai, Y.M. & Herman, J.P. Neural regulation of endocrine and autonomic stress responses. Nat. Rev. Neurosci. 10, 397–409 (2009).
Büdefeld, T., Grgurevic, N., Tobet, S.A. & Majdic, G. Sex differences in brain developing in the presence or absence of gonads. Dev. Neurobiol. 68, 981–995 (2008).
Tobet, S. et al. Brain sex differences and hormone influences: a moving experience? J. Neuroendocrinol. 21, 387–392 (2009).
Brown, A.E., Mani, S. & Tobet, S.A. The preoptic area/anterior hypothalamus of different strains of mice: sex differences and development. Brain Res. Dev. Brain Res. 115, 171–182 (1999).
Peinado, J.R. et al. Strain-dependent influences on the hypothalamo-pituitary-adrenal axis profoundly affect the 7B2 and PC2 null phenotypes. Endocrinology 146, 3438–3444 (2005).
Rinn, J.L. & Snyder, M. Sexual dimorphism in mammalian gene expression. Trends Genet. 21, 298–305 (2005).
Blackshaw, S. et al. Genomic analysis of mouse retinal development. PLoS Biol. 2, e247 (2004).
Chiang, C. et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413 (1996).
Kiecker, C. & Lumsden, A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat. Neurosci. 7, 1242–1249 (2004).
Kataoka, A. & Shimogori, T. Fgf8 controls regional identity in the developing thalamus. Development 135, 2873–2881 (2008).
Grove, E.A., Tole, S., Limon, J., Yip, L. & Ragsdale, C.W. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development 125, 2315–2325 (1998).
Herrera, E. et al. Foxd1 is required for proper formation of the optic chiasm. Development 131, 5727–5739 (2004).
Hatini, V., Tao, W. & Lai, E. Expression of winged helix genes, BF-1 and BF-2, define adjacent domains within the developing forebrain and retina. J. Neurobiol. 25, 1293–1309 (1994).
Pera, E.M. & Kessel, M. Demarcation of ventral territories by the homeobox gene NKX2.1 during early chick development. Dev. Genes Evol. 208, 168–171 (1998).
Miura, H., Yanazawa, M., Kato, K. & Kitamura, K. Expression of a novel aristaless related homeobox gene Arx in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 65, 99–109 (1997).
Watson, S.J., Barchas, J.D. & Li, C.H. beta-Lipotropin: localization of cells and axons in rat brain by immunocytochemistry. Proc. Natl. Acad. Sci. USA 74, 5155–5158 (1977).
Ino, H. Immunohistochemical characterization of the orphan nuclear receptor ROR alpha in the mouse nervous system. J. Histochem. Cytochem. 52, 311–323 (2004).
Moga, M.M. & Moore, R.Y. Organization of neural inputs to the suprachiasmatic nucleus in the rat. J. Comp. Neurol. 389, 508–534 (1997).
Fuller, P.M., Lu, J. & Saper, C.B. Differential rescue of light- and food-entrainable circadian rhythms. Science 320, 1074–1077 (2008).
Szabó, N.E. et al. Role of neuroepithelial Sonic hedgehog in hypothalamic patterning. J. Neurosci. 29, 6989–7002 (2009).
Dassule, H.R., Lewis, P., Bei, M., Maas, R. & McMahon, A.P. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775–4785 (2000).
Xu, Q., Tam, M. & Anderson, S.A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16–29 (2008).
Scholpp, S., Wolf, O., Brand, M. & Lumsden, A. Hedgehog signaling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development 133, 855–864 (2006).
Szabó, N.E., Zhao, T., Zhou, X. & Alvarez-Bolado, G. The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J. Neurosci. 29, 2453–2466 (2009).
Mathieu, J., Barth, A., Rosa, F.M., Wilson, S.W. & Peyrieras, N. Distinct and cooperative roles for Nodal and Hedgehog signals during hypothalamic development. Development 129, 3055–3065 (2002).
Tobet, S.A., Paredes, R.G., Chickering, T.W. & Baum, M.J. Telencephalic and diencephalic origin of radial glial processes in the developing preoptic area/anterior hypothalamus. J. Neurobiol. 26, 75–86 (1995).
Porteus, M.H. et al. DLX-2, MASH-1, and MAP-2 expression and bromodeoxyuridine incorporation define molecularly distinct cell populations in the embryonic mouse forebrain. J. Neurosci. 14, 6370–6383 (1994).
Hochberg, Y. & Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818 (1990).
Acknowledgements
We would like to thank C.-M. Fan for guidance concerning dissection of embryonic hypothalamus and S. Hattar, A. Moore and B. Nelson for comments on the manuscript. This work was supported by the RIKEN Brain Science Institute and Human Frontier Science Project (T.S.), a National Science Foundation Predoctoral Fellowship (D.A.L.), a Basil O'Connor Starter Scholar Award from the March of Dimes (S.B.) and an award from the Klingenstein Fund (S.B.). S.B. is a W.M. Keck Distinguished Young Investigator in Medical Science.
Author information
Authors and Affiliations
Contributions
S.B. conceived the study. L.Q. and L.J. dissected tissue, purified RNA and conducted microarray analysis. Y.Y. and J.Q. analyzed microarray data. D.A.L., A.M.-A., L.J., H.W. and M.A. carried out first-pass single-color ISH analysis. A.K., A.C.Y., H.M., L.J. and H.W. carried out the two-color ISH analysis. T.S. and S.B. contributed to the experimental design, supervised experiments, discussed the results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 5712 kb)
Supplementary Table 1
Normalized log(2) signal intensity is shown for all probe sets and all microarray samples examined in this study (XLS 29443 kb)
Supplementary Table 2
Median log(2) signal intensity is shown for all 25,471 probe sets that showed significantly different expression (P< 0.01 via one-way ANOVA analysis) at one or more time points when compared to median intensity for that probe set across all time points analyzed. (XLS 7210 kb)
Supplementary Table 3
Summary of all first-pass single color ISH data. (XLS 267 kb)
Supplementary Table 4
Summary of all two-color ISH data. (XLS 260 kb)
Rights and permissions
About this article
Cite this article
Shimogori, T., Lee, D., Miranda-Angulo, A. et al. A genomic atlas of mouse hypothalamic development. Nat Neurosci 13, 767–775 (2010). https://doi.org/10.1038/nn.2545
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2545
This article is cited by
-
Increased enhancer–promoter interactions during developmental enhancer activation in mammals
Nature Genetics (2024)
-
Transcriptomic survey of key reproductive and metabolic tissues in mouse models of polycystic ovary syndrome
Communications Biology (2023)
-
Dorsoventral Arrangement of Lateral Hypothalamus Populations in the Mouse Hypothalamus: a Prosomeric Genoarchitectonic Analysis
Molecular Neurobiology (2023)
-
Populational heterogeneity and partial migratory origin of the ventromedial hypothalamic nucleus: genoarchitectonic analysis in the mouse
Brain Structure and Function (2023)
-
Ontogenetic rules for the molecular diversification of hypothalamic neurons
Nature Reviews Neuroscience (2022)