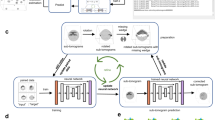
Abstract
Cellular electron cryotomography offers researchers the ability to observe macromolecules frozen in action in situ, but a primary challenge with this technique is identifying molecular components within the crowded cellular environment. We introduce a method that uses neural networks to dramatically reduce the time and human effort required for subcellular annotation and feature extraction. Subsequent subtomogram classification and averaging yield in situ structures of molecular components of interest. The method is available in the EMAN2.2 software package.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Lučič, V., Rigort, A. & Baumeister, W. J. Cell Biol. 202, 407–419 (2013).
Galaz-Montoya, J.G. et al. J. Struct. Biol. 194, 383–394 (2016).
Chen, Y., Pfeffer, S., Hrabe, T., Schuller, J.M. & Förster, F. J. Struct. Biol. 182, 235–245 (2013).
Asano, S. et al. Science 347, 439–442 (2015).
Pfeffer, S., Woellhaf, M.W., Herrmann, J.M. & Förster, F. Nat. Commun. 6, 6019 (2015).
Ding, H.J., Oikonomou, C.M. & Jensen, G.J. J. Struct. Biol. 192, 279–286 (2015).
Rigort, A. et al. J. Struct. Biol. 177, 135–144 (2012).
Page, C., Hanein, D. & Volkmann, N. Ultramicroscopy 155, 20–26 (2015).
Frangakis, A.S. et al. Proc. Natl. Acad. Sci. USA 99, 14153–14158 (2002).
LeCun, Y., Bengio, Y. & Hinton, G. Nature 521, 436–444 (2015).
Garduño, E., Wong-Barnum, M., Volkmann, N. & Ellisman, M.H. J. Struct. Biol. 162, 368–379 (2008).
Hecksel, C.W. et al. Microsc. Microanal. 22, 487–496 (2016).
Wang, R. et al. Proc. Natl. Acad. Sci. USA 112, 14266–14271 (2015).
Dai, W. et al. Nature 502, 707–710 (2013).
Hashem, Y. et al. Nature 494, 385–389 (2013).
Asenjo, A.B. et al. Cell Rep. 3, 759–768 (2013).
Koning, R.I. et al. J. Struct. Biol. 161, 459–468 (2008).
Garvalov, B.K. et al. J. Cell Biol. 174, 759–765 (2006).
Scheuring, S. & Sturgis, J.N. Science 309, 484–487 (2005).
Tang, G. et al. J. Struct. Biol. 157, 38–46 (2007).
Nair, V. & Hinton, G.E. In Proc. 27th Int. Conf. Mach. Learn. (eds. Fürnkranz, J. & Joachims, T.) 807–814 (ICML, 2010).
Vincent, P., Larochelle, H., Bengio, Y. & Manzagol, P.-A. In Proc. 25th Int. Conf. Mach. Learn (eds. McCallum, A. & Roweis, S.) 1096–1103 (ICML, 2008).
Hinton, G.E., Srivastava, N., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R.R. Preprint at http://arxiv.org/abs/1207.0580 (2012).
Dieleman, S., Willett, K.W. & Dambre, J. Mon. Not. R. Astron. Soc. 450, 1441–1459 (2015).
Zhou, J. & Troyanskaya, O.G. Nat. Methods 12, 931–934 (2015).
Noh, H., Hong, S. & Han, B. Preprint at http://arxiv.org/abs/1505.04366 (2015).
Krizhevsky, A., Sutskever, I. & Hinton, G.E. In Advances in Neural Information Processing Systems 25 (eds. Pereira, F., Burges, C.J.C., Bottou, L. & Weinberger, K.Q.) 1097–1105 (Curran Associates, 2012).
Dai, W. et al. Nat. Protoc. 9, 2630–2642 (2014).
Apostol, B.L. et al. Proc. Natl. Acad. Sci. USA 100, 5950–5955 (2003).
Wirtz, E., Leal, S., Ochatt, C. & Cross, G.A. Mol. Biochem. Parasitol. 99, 89–101 (1999).
Kaminsky, R., Beaudoin, E. & Cunningham, I. Acta Trop. 45, 33–43 (1988).
Chen, M. et al. Protocol Exchange https://doi.org/10.1038/nprot.2017.095 (2017).
Acknowledgements
We gratefully acknowledge support of NIH grants (R01GM080139, P01NS092525, P41GM103832), Ovarian Cancer Research Fund and Singapore Ministry of Education. Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Author information
Authors and Affiliations
Contributions
M.C. designed the protocol. W.D., S.Y.S. and C.Y.H. provided the test data sets. M.C. and D.J. tested and refined the protocol. M.C., W.D., S.Y.S., M.F.S., W.C. and S.J.L. wrote the paper and provided suggestions during development.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Missing wedge artifact in CryoET.
a. Slice view of a tomogram in the X-Y, Y-Z and X-Z plane. b. Fourier Transform of projection of the same tomogram in X-Y, Y-Z and X-Z plane.
Supplementary Figure 3 Reusability of trained neural network.
a-d. Annotation of ribosomes in a tomogram of cyanobacteria taken with a Zernike phase plate using different CNNs. a. Ribosomes annotated by a CNN trained on 5 positive and 50 negative samples from the same tomogram (89% correct). b. Using a CNN trained on 5 positive and 50 negative samples from another ZPP tomogram (86% correct). c. Using a CNN trained on 5 positive and 50 negative samples from a non ZPP tomogram (78% correct). d. Using a CNN trained on 10 positive and 100 negative samples from the tomogram in a (91.3% correct). e-h. Annotation of microtubules in two tomograms (I and II) of PC12 cells using two CNNs. CNN A is trained on 5 positive and 50 negative samples from tomogram I. CNN B is trained on 5 positive and 50 negative samples from tomogram II. e. Microtubules in tomogram I annotated by CNN A. f. Microtubules in tomogram II annotated by CNN A. g. Microtubules in tomogram I annotated by CNN B. h. Microtubules in tomogram II annotated by CNN B.
Supplementary Figure 4 Comparison of raw tomogram slices and corresponding automatic annotation in Fig 2.
Supplementary Figure 5 Impact of training inaccuracy on CNN annotations.
a. One of the training samples of microtubules of PC12 cell used in Fig.1b. b. Corresponding manual annotation with a minor error marked by the red arrow. c. Annotation of the patch using a CNN trained on the sample with incorrect annotation and all the other positive and negative samples with correct manual annotation used in Fig.1b. Note the error is fixed in the annotation. d. A slice view of the tomogram. e. Annotation of the slice using the CNN in c.
Supplementary Figure 6 Removal of high contrast artifact.
a. Volume rendering of a region of the tomogram shown in Fig 1. b. Annotation of double membrane in the tomogram using the same CNN used in Fig 1. Note the carbon edge is falsely recognized as double membrane. c. Automatic annotation of carbon edge using a CNN. d. Removal of carbon edge from double membrane annotation in b.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1.
Supplementary Protocol
Supplementary Protocol.
Workflow of automated tomogram annotation
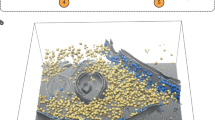
Demonstration of tomogram annotation workflow with our software package, using a PC12 cell tomogram as an example. The process includes selection of training set, annotation of the tomogram, and subtomogram averaging results.
Annotation of the PC12 cell tomogram
Movie showing the PC12 cell tomogram used in the figure and its annotation, including volume rendering and slice view of the input tomogram, and a 3D view of annotated features.
Annotation of the human platelet cell tomogram
Movie showing the human platelet cell tomogram used in the figure and its annotation, including volume rendering and slice view of the input tomogram, and a 3D view of annotated features.
Annotation of the African Trypanosomes cell tomogram
Movie showing the African Trypanosomes cell tomogram used in the figure and its annotation, including volume rendering and slice view of the input tomogram, and a 3D view of annotated features.
Annotation of the Cyanobacteria cell tomogram
Movie showing the Cyanobacteria cell tomogram used in the figure and its annotation, including volume rendering and slice view of the input tomogram, and a 3D view of annotated features.
Rights and permissions
About this article
Cite this article
Chen, M., Dai, W., Sun, S. et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat Methods 14, 983–985 (2017). https://doi.org/10.1038/nmeth.4405
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4405
This article is cited by
-
Electrospray-assisted cryo-EM sample preparation to mitigate interfacial effects
Nature Methods (2024)
-
Ultrastructure of macromolecular assemblies contributing to bacterial spore resistance revealed by in situ cryo-electron tomography
Nature Communications (2024)
-
Vimentin filaments integrate low-complexity domains in a complex helical structure
Nature Structural & Molecular Biology (2024)
-
Modular characterization of SARS-CoV-2 nucleocapsid protein domain functions in nucleocapsid-like assembly
Molecular Biomedicine (2023)
-
Previously uncharacterized rectangular bacterial structures in the dolphin mouth
Nature Communications (2023)