Abstract
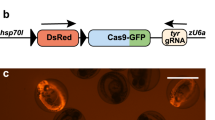
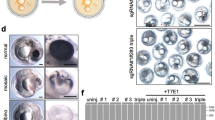
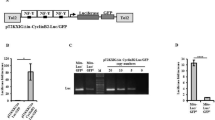
To trace cell lineages in a developing vertebrate and to observe, in vivo, how behaviors of individual cells are affected by the genes they express, we created a zebrafish line containing a transgene called mosaic analysis in zebrafish (MAZe), built around a self-excising hsp70:Cre cassette. Heat shock triggers Cre recombinase–mediated recombination in a random subset of cells, bringing the transcriptional activator Gal4:VP16 under control of the EF1α promoter. Gal4-VP16 then activates expression of a fluorescent protein from an upstream activating sequence (UAS) promoter. Marked clones of cells expressing any desired gene product can be generated by crossing MAZe fish with other lines containing UAS-driven transgenes. The number of clones induced, and their time of origin, could be varied by adjusting heat-shock timing and duration. As an alternative to heat shock, we introduced Cre under a tissue-specific promoter in MAZe fish to generate clones in a designated tissue.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Stern, C. Somatic crossing over and segregation in Drosophila melanogaster . Genetics 21, 625–730 (1936).
Stern, C. Genetic mosaics, and other essays (Harvard University Press, Cambridge, Massachusetts, USA, 1968).
Le Douarin, N.M. & Kalcheim, C. The Neural Crest (Cambridge University Press, New York, 1999).
McLaren, A. Mammalian Chimaeras (Cambridge University Press, Cambridge, 1976).
Xu, T. & Rubin, G.M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237 (1993).
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254 (2001).
Duffy, J.B. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34, 1–15 (2002).
Branda, C.S. & Dymecki, S.M. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 (2004).
Westerfield, M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) (University of Oregon Press, Eugene, Oregon, USA, 2000).
Halloran, M.C. et al. Laser-induced gene expression in specific cells of transgenic zebrafish. Development 127, 1953–1960 (2000).
Hans, S. et al. Temporally-controlled site-specific recombination in zebrafish. PLoS One 4, e4640 (2009).
Halpern, M.E. et al. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97–110 (2008).
Scheer, N. & Campos-Ortega, J.A. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech. Dev. 80, 153–158 (1999).
Lewis, J. Notch signalling and the control of cell fate choices in vertebrates. Semin. Cell Dev. Biol. 9, 583–589 (1998).
Kim, C.H. et al. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216, 109–112 (1996).
Offield, M.F., Hirsch, N. & Grainger, R.M. The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development 127, 1789–1797 (2000).
Dutton, J.R. et al. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev. Biol. 8, 105 (2008).
Hardy, M.E., Ross, L.V. & Chien, C.B. Focal gene misexpression in zebrafish embryos induced by local heat shock using a modified soldering iron. Dev. Dyn. 236, 3071–3076 (2007).
Urasaki, A., Morvan, G. & Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 (2006).
Bunting, M. et al. Targeting genes for self-excision in the germ line. Genes Dev. 13, 1524–1528 (1999).
Grabher, C. & Wittbrodt, J. Efficient activation of gene expression using a heat-shock inducible Gal4/Vp16-UAS system in medaka. BMC Biotechnol. 4, 26 (2004).
Shaner, N.C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
Okada, A. et al. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp. Neurol. 156, 394–406 (1999).
Tawk, M. et al. A mirror-symmetric cell division that orch-estrates neuroepithelial morphogenesis. Nature 446, 797–800 (2007).
Kajita, M. et al. Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Srctransformed cells. J. Cell Sci. 123, 171–180 (2010).
Grabher, C., Joly, J.S. & Wittbrodt, J. Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI. Methods Cell Biol. 77, 381–401 (2004).
Acknowledgements
We thank M. Tada (University College London) for the UAS:gfp transgenic line; R. Kelsh (University of Bath) for the Sox10:gfp transgenic line and the Sox10 promoter sequence; J. Clarke (Kings College London) for the Gap42-GFP plasmid; M. Capecchi (University of Utah School of Medicine) for the pACN plasmid; H. Gerhardt (London Research Institute, Cancer Research UK) for the Cre antibody; L. Zimmerman (National Institute for Medical Research, London) for the γ-crystallin promoter; P. Taylor and D. Martin for fish husbandry; J. Clarke, S. Wilson and D. Ish-Horowicz for comments on the manuscript; and Cancer Research UK for funding.
Author information
Authors and Affiliations
Contributions
R.T.C. conceived, designed and generated the MAZe transgenic fish; R.T.C. and C.L. characterized the MAZe line; and R.T.C., C.L. and J.L. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2 (PDF 7981 kb)
Supplementary Video 1
Myoblast fusion (MAZe + membrane GFP). A MAZe-labeled myoblast (left red arrow) fuses with neighboring unlabeled myoblast in the dorsal part of a trunk somite. The unlabeled myoblast nucleus becomes fluorescent after cytoplasm continuity is observed (yellow arrow). Thereafter, both nuclei are fluorescent (red arrows) and the intensity of their fluorescence increases over time. MAZe embryos were injected with 100 pg of mRNA encoding membrane-targeted GFP at the 1-2 cell stage and heat-shocked at 39°C for 45 min at 8 h.p.f.; confocal images were obtained from ~18 h.p.f. every 14 min. Each time point corresponds to a single optical section of 3 μm depth. Anterior to the left, dorsal up. (MOV 1667 kb)
Supplementary Video 2
Myoblast fusion (MAZe + UAS:gfp). A myoblast labeled with MAZe and GFP (red arrow) in the medial part of a trunk somite fuses with a neighboring unlabeled myoblast. The myoblast cytoplasm (observed with GFP fluorescence) elongates to span the entire length of the somite, and subsequently a second fluorescent nucleus (second red arrow) is observed in the same cell. Embryos obtained from crossing MAZe to UAS:gfp fish were heat shocked at 39°C for 45 min at 8 h.p.f. and confocal images were obtained from ~18 h.p.f. every 14 min. Each time point corresponds to a single optical section of 3 μm depth. (MOV 1031 kb)
Supplementary Video 3
Dividing MAZe-labeled cell. A MAZe-labeled cell situated just dorsal to the notochord (white arrow) divides. As the nuclear membrane breaks down the fluorescent protein diffuses to fill the cytoplasm of the cell. After cytokinesis the nlsRFP becomes re-localized to the nuclei of the daughter cells, labeling each nucleus with equal intensity. MAZe embryos were heat shocked at 39°C for 45 min at 8 h.p.f. and confocal and bright-field images were obtained from ~30 h.p.f. every 8 min. Each time point corresponds to a projection of confocal sections merged with the corresponding bright field image. Anterior to the left, dorsal up. (MOV 909 kb)
Rights and permissions
About this article
Cite this article
Collins, R., Linker, C. & Lewis, J. MAZe: a tool for mosaic analysis of gene function in zebrafish. Nat Methods 7, 219–223 (2010). https://doi.org/10.1038/nmeth.1423
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1423
This article is cited by
-
Aurora kinase B regulates axonal outgrowth and regeneration in the spinal motor neurons of developing zebrafish
Cellular and Molecular Life Sciences (2018)
-
Genetics of photoreceptor degeneration and regeneration in zebrafish
Cellular and Molecular Life Sciences (2011)
-
In vivo imaging of hematopoietic stem cell development in the zebrafish
Frontiers of Medicine (2011)
-
aMAZe-ing tools for mosaic analysis in zebrafish
Nature Methods (2010)