Abstract
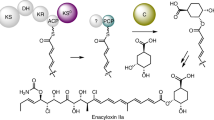
Ketosynthases produce the carbon backbones of a vast number of biologically active polyketides by catalyzing Claisen condensations of activated acyl and malonyl building blocks. Here we report that a ketosynthase homolog from Streptomyces tendae, CerJ, unexpectedly forms malonyl esters during the biosynthesis of cervimycin, a glycoside antibiotic against methicillin-resistant Staphylococcus aureus (MRSA). Deletion of cerJ yielded a substantially more active cervimycin variant lacking the malonyl side chain, and in vitro biotransformations revealed that CerJ is capable of transferring malonyl, methylmalonyl and dimethylmalonyl units onto the glycoside. According to phylogenetic analyses and elucidation of the crystal structure, CerJ is functionally and structurally positioned between the ketosynthase catalyzing Claisen condensations and acyl-ACP shuttles, and it features a noncanonical catalytic triad. Site-directed mutagenesis and structures of CerJ in complex with substrates not only allowed us to establish a model for the reaction mechanism but also provided insights into the evolution of this important subclass of the thiolase superfamily.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 48, 4688–4716 (2009).
Staunton, J. & Weissman, K.J. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18, 380–416 (2001).
Rix, U., Fischer, C., Remsing, L.L. & Rohr, J. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep. 19, 542–580 (2002).
Khosla, C. Structures and mechanisms of polyketide synthases. J. Org. Chem. 74, 6416–6420 (2009).
Fischbach, M.A. & Walsh, C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006).
Hertweck, C., Luzhetskyy, A., Rebets, Y. & Bechthold, A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24, 162–190 (2007).
Austin, M.B. & Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110 (2003).
Tsay, J.T., Oh, W., Larson, T.J., Jackowski, S. & Rock, C.O. Isolation and characterization of the β-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J. Biol. Chem. 267, 6807–6814 (1992).
Pan, H. et al. Crystal structure of the priming β-ketosynthase from the R1128 polyketide biosynthetic pathway. Structure 10, 1559–1568 (2002).
Heath, R.J. & Rock, C.O. The claisen condensation in biology. Nat. Prod. Rep. 19, 581–596 (2002).
Herold, K. et al. Cervimycin A-D: a polyketide glycoside complex from a cave bacterium can defeat vancomycin resistance. Chem. Eur. J. 11, 5523–5530 (2005).
Herold, K., Xu, Z.L., Gollmick, F.A., Gräfe, U. & Hertweck, C. Biosynthesis of cervimycin C, an aromatic polyketide antibiotic bearing an unusual dimethylmalonyl moiety. Org. Biomol. Chem. 2, 2411–2414 (2004).
Moore, B.S. & Hertweck, C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 19, 70–99 (2002).
Bao, W., Sheldon, P.J. & Hutchinson, C.R. Purification and properties of the Streptomyces peucetius DpsC β-ketoacyl:acyl carrier protein synthase III that specifies the propionate-starter unit for type II polyketide biosynthesis. Biochemistry 38, 9752–9757 (1999).
Bao, W., Sheldon, P.J., Wendt-Pienkowski, E. & Hutchinson, C.R. The Streptomyces peucetius dpsC gene determines the choice of starter unit in biosynthesis of the daunorubicin polyketide. J. Bacteriol. 181, 4690–4695 (1999).
Möllmann, U. et al. HKI10311129, neues Antibiotikum, Verfahren zu dessen Herstellung und Arzneimittel enthaltend HKI10311129. German patent vol. DE 10 2004 010 219 B4 2007.02.01 (2004).
Neises, B. & Steglich, W. Simple method for the esterification of carboxylic acids. Angew. Chem. Int. Edn Engl. 17, 522–524 (1978).
Mathieu, M. et al. The 2.8 Å crystal structure of peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: a five-layered αβαβα structure constructed from two core domains of identical topology. Structure 2, 797–808 (1994).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Davies, C., Heath, R.J., White, S.W. & Rock, C.O. The 1.8 Å crystal structure and active-site architecture of β-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure 8, 185–195 (2000).
Gajiwala, K.S. et al. Crystal structures of bacterial FabH suggest a molecular basis for the substrate specificity of the enzyme. FEBS Lett. 583, 2939–2946 (2009).
Oke, M. et al. The Scottish Structural Proteomics Facility: targets, methods and outputs. J. Struct. Funct. Genomics 11, 167–180 (2010).
Qiu, X. et al. Crystal structure and substrate specificity of the β-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Protein Sci. 14, 2087–2094 (2005).
Brown, A.K. et al. Probing the mechanism of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein synthase III mtFabH: factors influencing catalysis and substrate specificity. J. Biol. Chem. 280, 32539–32547 (2005).
Xu, Z., Schenk, A. & Hertweck, C. Molecular analysis of the benastatin biosynthetic pathway and genetic engineering of altered fatty acid-polyketide hybrids. J. Am. Chem. Soc. 129, 6022–6030 (2007).
Xu, Z., Metsä-Ketelä, M. & Hertweck, C. Ketosynthase III as a gateway to engineering the biosynthesis of antitumoral benastatin derivatives. J. Biotechnol. 140, 107–113 (2009).
Bera, A.K. et al. Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate. Biochemistry 48, 8644–8655 (2009).
White, S.W., Zheng, J., Zhang, Y.M. & Rock The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 (2005).
Qiu, X. et al. Refined structures of β-ketoacyl-acyl carrier protein synthase III. J. Mol. Biol. 307, 341–356 (2001).
He, Q.L. et al. Dissection of two acyl-transfer reactions centered on acyl-S-carrier protein intermediates for incorporating 5-chloro-6-methyl-O-methylsalicyclic acid into chlorothricin. ChemBioChem 10, 813–819 (2009).
Serre, L., Verbree, E.C., Dauter, Z., Stuitje, A.R. & Derewenda, Z.S. The Escherichia coli malonyl-CoA:acyl carrier protein transacylase at 1.5-Å resolution. Crystal structure of a fatty acid synthase component. J. Biol. Chem. 270, 12961–12964 (1995).
Pojer, F., Li, S.M. & Heide, L. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148, 3901–3911 (2002).
Wang, Z.X., Li, S.M. & Heide, L. Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob. Agents Chemother. 44, 3040–3048 (2000).
Dürr, C. et al. Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tu6071: Analysis of the gene cluster and generation of derivatives. Chem. Biol. 13, 365–377 (2006).
Daum, M. et al. Organisation of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. ChemBioChem 10, 1073–1083 (2009).
Jia, X.Y. et al. Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem. Biol. 13, 575–585 (2006).
Weitnauer, G. et al. Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tu57 and production of new antibiotics. Chem. Biol. 8, 569–581 (2001).
Ahlert, J. et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297, 1173–1176 (2002).
Anderle, C. et al. Biosynthesis of clorobiocin: investigation of the transfer and methylation of the pyrrolyl-2-carboxyl moiety. Arch. Microbiol. 187, 227–237 (2007).
Balibar, C.J., Garneau-Tsodikova, S. & Walsh, C.T. Covalent CouN7 enzyme intermediate for acyl group shuttling in aminocoumarin biosynthesis. Chem. Biol. 14, 679–690 (2007).
Strieter, E.R., Vaillancourt, F.H. & Walsh, C.T. CmaE: a transferase shuttling aminoacyl groups between carrier protein domains in the coronamic acid biosynthetic pathway. Biochemistry 46, 7549–7557 (2007).
Fullone, M.R. et al. Mutational analysis and homology modelling of SyrC, the aminoacyltransferase in the biosynthesis of syringomycin. Biochem. Biophys. Res. Commun. 364, 201–207 (2007).
Freitag, A., Wemakor, E., Li, S.M. & Heide, L. Acyl transfer in clorobiocin biosynthesis: involvement of several proteins in the transfer of the pyrrole-2-carboxyl moiety to the deoxysugar. ChemBioChem 6, 2316–2325 (2005).
Kwon, H.J. et al. C-O bond formation by polyketide synthases. Science 297, 1327–1330 (2002).
Haapalainen, A.M., Merilainen, G. & Wierenga, R.K. The thiolase superfamily: condensing enzymes with diverse reaction specificities. Trends Biochem. Sci. 31, 64–71 (2006).
Igual, J.C., Gonzalez-Bosch, C., Dopazo, J. & Perez-Ortin, J.E. Phylogenetic analysis of the thiolase family. Implications for the evolutionary origin of peroxisomes. J. Mol. Evol. 35, 147–155 (1992).
Acknowledgements
We thank the German Federal Ministry of Education and Research (GenoMik) for financial support and the Swiss light source beamline X06DA for offering beamtime.
Author information
Authors and Affiliations
Contributions
T.B. and M.U. performed biochemical and genetic experiments as well as bioinformatic analysis. K.S. performed chemical analysis. G.Z. and T.S. performed crystallographic studies. G.Z. and C.H. designed research. G.Z., T.B. and C.H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 3552 kb)
Rights and permissions
About this article
Cite this article
Bretschneider, T., Zocher, G., Unger, M. et al. A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis. Nat Chem Biol 8, 154–161 (2012). https://doi.org/10.1038/nchembio.746
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.746
This article is cited by
-
No job too small for a giant enzyme
Nature Chemical Biology (2023)
-
Integrated omics unveil the secondary metabolic landscape of a basal dinoflagellate
BMC Biology (2020)
-
Dual phenazine gene clusters enable diversification during biosynthesis
Nature Chemical Biology (2019)
-
Polyketide synthase chimeras reveal key role of ketosynthase domain in chain branching
Nature Chemical Biology (2015)
-
Biosynthesis and function of bacterial dialkylresorcinol compounds
Applied Microbiology and Biotechnology (2015)