Abstract
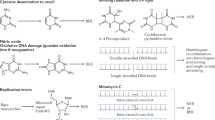
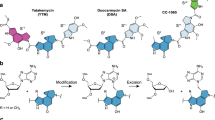
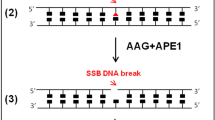
Yatakemycin (YTM) is an extraordinarily toxic DNA alkylating agent with potent antimicrobial and antitumor properties and is the most recent addition to the CC-1065 and duocarmycin family of natural products. Though bulky DNA lesions the size of those produced by YTM are normally removed from the genome by the nucleotide-excision repair (NER) pathway, YTM adducts are also a substrate for the bacterial DNA glycosylases AlkD and YtkR2, unexpectedly implicating base-excision repair (BER) in their elimination. The reason for the extreme toxicity of these lesions and the molecular basis for the way they are eliminated by BER have been unclear. Here, we describe the structural and biochemical properties of YTM adducts that are responsible for their toxicity, and define the mechanism by which they are excised by AlkD. These findings delineate an alternative strategy for repair of bulky DNA damage and establish the cellular utility of this pathway relative to that of NER.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Friedberg, E.C. et al. DNA Repair and Mutagenesis (ASM Press, 2006).
Truglio, J.J., Croteau, D.L., Van Houten, B. & Kisker, C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106, 233–252 (2006).
Nouspikel, T. DNA repair in mammalian cells: nucleotide excision repair: variations on versatility. Cell. Mol. Life Sci. 66, 994–1009 (2009).
Fromme, J.C. & Verdine, G.L. Base excision repair. Adv. Protein Chem. 69, 1–41 (2004).
Hitomi, K., Iwai, S. & Tainer, J.A. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst.) 6, 410–428 (2007).
Brooks, S.C., Adhikary, S., Rubinson, E.H. & Eichman, B.F. Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta 1834, 247–271 (2013).
Slupphaug, G. et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384, 87–92 (1996).
Stivers, J.T. Site-specific DNA damage recognition by enzyme-induced base flipping. Prog. Nucleic Acid Res. Mol. Biol. 77, 37–65 (2004).
Mullins, E.A. et al. The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions. Nature 527, 254–258 (2015).
Rubinson, E.H., Gowda, A.S., Spratt, T.E., Gold, B. & Eichman, B.F. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature 468, 406–411 (2010).
Hanka, L.J., Dietz, A., Gerpheide, S.A., Kuentzel, S.L. & Martin, D.G. CC-1065 (NSC-298223), a new antitumor antibiotic. Production, in vitro biological activity, microbiological assays and taxonomy of the producing microorganism. J. Antibiot. (Tokyo) 31, 1211–1217 (1978).
Takahashi, I. et al. Duocarmycin A, a new antitumor antibiotic from Streptomyces. J. Antibiot. (Tokyo) 41, 1915–1917 (1988).
Ichimura, M. et al. Duocarmycin SA, a new antitumor antibiotic from Streptomyces sp. J. Antibiot. (Tokyo) 43, 1037–1038 (1990).
Igarashi, Y. et al. Yatakemycin, a novel antifungal antibiotic produced by Streptomyces sp. TP-A0356. J. Antibiot. (Tokyo) 56, 107–113 (2003).
Xu, H. et al. Self-resistance to an antitumor antibiotic: a DNA glycosylase triggers the base-excision repair system in yatakemycin biosynthesis. Angew. Chem. Int. Ed. Engl. 51, 10532–10536 (2012).
Martin, D.G. et al. CC-1065 (NSC 298223), a potent new antitumor agent: improved production and isolation, characterization and antitumor activity. J. Antibiot. (Tokyo) 34, 1119–1125 (1981).
Lin, C.H. & Patel, D.J. Solution structure of the covalent duocarmycin A-DNA duplex complex. J. Mol. Biol. 248, 162–179 (1995).
Eis, P.S. et al. High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA. J. Mol. Biol. 272, 237–252 (1997).
Schnell, J.R., Ketchem, R.R., Boger, D.L. & Chazin, W.J. Binding-induced activation of DNA alkylation by duocarmycin SA: insights from the structure of an indole derivative-DNA adduct. J. Am. Chem. Soc. 121, 5645–5652 (1999).
Parrish, J.P., Kastrinsky, D.B., Wolkenberg, S.E., Igarashi, Y. & Boger, D.L. DNA alkylation properties of yatakemycin. J. Am. Chem. Soc. 125, 10971–10976 (2003).
Tichenor, M.S. et al. Systematic exploration of the structural features of yatakemycin impacting DNA alkylation and biological activity. J. Am. Chem. Soc. 129, 10858–10869 (2007).
Swenson, D.H. et al. Mechanism of interaction of CC-1065 (NSC 298223) with DNA. Cancer Res. 42, 2821–2828 (1982).
Gunz, D., Hess, M.T. & Naegeli, H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 271, 25089–25098 (1996).
Selby, C.P. & Sancar, A. ABC excinuclease incises both 5′ and 3′ to the CC-1065-DNA adduct and its incision activity is stimulated by DNA helicase II and DNA polymerase I. Biochemistry 27, 7184–7188 (1988).
Jin, S.G. et al. Excision repair of adozelesin-N3 adenine adduct by 3-methyladenine-DNA glycosylases and UvrABC nuclease. Mol. Cells 11, 41–47 (2001).
Kiakos, K. et al. DNA sequence selective adenine alkylation, mechanism of adduct repair, and in vivo antitumor activity of the novel achiral seco-amino-cyclopropylbenz[e]indolone analogue of duocarmycin AS-I-145. Mol. Cancer Ther. 6, 2708–2718 (2007).
Gray, D.M., Ratliff, R.L. & Vaughan, M.R. Circular dichroism spectroscopy of DNA. Methods Enzymol. 211, 389–406 (1992).
Franklin, R.E. & Gosling, R.G. Molecular configuration in sodium thymonucleate. Nature 171, 740–741 (1953).
Lucas, A.A., Lambin, P., Mairesse, R. & Mathot, M. Revealing the backbone structure of B-DNA from laser optical simulations of its X-ray diffraction diagram. J. Chem. Educ. 76, 378–383 (1999).
Lucas, A.A. A-DNA and B-DNA: comparing their historical X-ray fiber diffraction images. J. Chem. Educ. 85, 737–743 (2008).
Shibasaki, K., Fujii, A., Mikami, N. & Tsuzuki, S. Magnitude of the CH/π interaction in the gas phase: experimental and theoretical determination of the accurate interaction energy in benzene-methane. J. Phys. Chem. A 110, 4397–4404 (2006).
Osborne, M.R. & Phillips, D.H. Preparation of a methylated DNA standard, and its stability on storage. Chem. Res. Toxicol. 13, 257–261 (2000).
Parsons, Z.D., Bland, J.M., Mullins, E.A. & Eichman, B.F. A catalytic role for C-H/π interactions in base excision repair by Bacillus cereus DNA glycosylase AlkD. J. Am. Chem. Soc. 138, 11485–11488 (2016).
Ramstein, J. & Lavery, R. Energetic coupling between DNA bending and base pair opening. Proc. Natl. Acad. Sci. USA 85, 7231–7235 (1988).
Yang, W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair (Amst.) 5, 654–666 (2006).
Gold, B., Stone, M.P. & Marky, L.A. Looking for Waldo: a potential thermodynamic signature to DNA damage. Acc. Chem. Res. 47, 1446–1454 (2014).
Mullins, E.A., Rubinson, E.H. & Eichman, B.F. The substrate binding interface of alkylpurine DNA glycosylase AlkD. DNA Repair (Amst.) 13, 50–54 (2014).
Barrett, T.E. et al. Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J. 18, 6599–6609 (1999).
Lee, S., Bowman, B.R., Ueno, Y., Wang, S. & Verdine, G.L. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J. Am. Chem. Soc. 130, 11570–11571 (2008).
Pidugu, L.S. et al. Structural basis for excision of 5-formylcytosine by thymine DNA glycosylase. Biochemistry 55, 6205–6208 (2016).
Stivers, J.T., Pankiewicz, K.W. & Watanabe, K.A. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry 38, 952–963 (1999).
Schärer, O.D., Kawate, T., Gallinari, P., Jiricny, J. & Verdine, G.L. Investigation of the mechanisms of DNA binding of the human G/T glycosylase using designed inhibitors. Proc. Natl. Acad. Sci. USA 94, 4878–4883 (1997).
Daley, J.M., Zakaria, C. & Ramotar, D. The endonuclease IV family of apurinic/apyrimidinic endonucleases. Mutat. Res. 705, 217–227 (2010).
Mullins, E.A., Shi, R., Kotsch, L.A. & Eichman, B.F. A new family of HEAT-like repeat proteins lacking a critical substrate recognition motif present in related DNA glycosylases. PLoS One 10, e0127733 (2015).
Noll, D.M., Mason, T.M. & Miller, P.S. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 106, 277–301 (2006).
Alseth, I. et al. A new protein superfamily includes two novel 3-methyladenine DNA glycosylases from Bacillus cereus, AlkC and AlkD. Mol. Microbiol. 59, 1602–1609 (2006).
Chen, Y.C., Li, C.L., Hsiao, Y.Y., Duh, Y. & Yuan, H.S. Structure and function of TatD exonuclease in DNA repair. Nucleic Acids Res. 42, 10776–10785 (2014).
Komaki, H., Ichikawa, N., Hosoyama, A., Fujita, N. & Igarashi, Y. Draft genome sequence of Streptomyces sp. TP-A0356, a producer of yatakemycin. Genome Announc. 3, e01446–15 (2015).
Wang, S. et al. Characterization of a novel DNA glycosylase from S. sahachiroi involved in the reduction and repair of azinomycin B induced DNA damage. Nucleic Acids Res. 44, 187–197 (2016).
Semlow, D.R., Zhang, J., Budzowska, M., Drohat, A.C. & Walter, J.C. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167, 498–511.e14 (2016).
Rubinson, E.H., Metz, A.H., O' Quin, J. & Eichman, B.F. A new protein architecture for processing alkylation damaged DNA: the crystal structure of DNA glycosylase AlkD. J. Mol. Biol. 381, 13–23 (2008).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Mullins, E.A. et al. An HPLC-tandem mass spectrometry method for simultaneous detection of alkylated base excision repair products. Methods 64, 59–66 (2013).
Acknowledgements
We thank Y. Igarashi (Toyama Prefectural University) for providing yatakemycin. This work was funded by the National Science Foundation (MCB-1517695) and the National Institutes of Health (R01 ES019625). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy Office of Science by Argonne National Laboratory, was supported by the US Department of Energy (DE-AC02-06CH11357). Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (085P1000817). E.A.M. was partially supported by the Vanderbilt Training Program in Environmental Toxicology (T32 ES007028).
Author information
Authors and Affiliations
Contributions
E.A.M. and B.F.E. conceived the project; E.A.M., R.S., and B.F.E. designed experiments; E.A.M. performed biochemical, biophysical, and structural experiments; R.S. performed cellular experiments; E.A.M., R.S., and B.F.E. analyzed data; E.A.M. and B.F.E. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–8 and Supplementary Tables 1–3 (PDF 13733 kb)
Rights and permissions
About this article
Cite this article
Mullins, E., Shi, R. & Eichman, B. Toxicity and repair of DNA adducts produced by the natural product yatakemycin. Nat Chem Biol 13, 1002–1008 (2017). https://doi.org/10.1038/nchembio.2439
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2439
This article is cited by
-
Bacterial DNA excision repair pathways
Nature Reviews Microbiology (2022)
-
Structural evolution of a DNA repair self-resistance mechanism targeting genotoxic secondary metabolites
Nature Communications (2021)
-
Non-flipping DNA glycosylase AlkD scans DNA without formation of a stable interrogation complex
Communications Biology (2021)
-
Insights into conformational changes in AlkD bound to DNA with a yatakemycin adduct from computational simulations
Theoretical Chemistry Accounts (2018)