Abstract
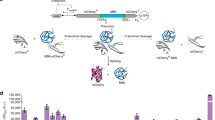
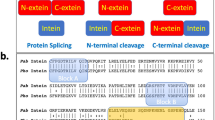
Protein sequences are diversified on the DNA level by recombination and mutation and can be further increased on the RNA level by alternative RNA splicing, involving introns that have important roles in many biological processes. The protein version of introns (inteins), which catalyze protein splicing, were first reported in the 1990s. The biological roles of protein splicing still remain elusive because inteins neither provide any clear benefits nor have an essential role in their host organisms. We now report protein alternative splicing, in which new protein sequences can be produced by protein recombination by intermolecular domain swapping of inteins, as elucidated by NMR spectroscopy and crystal structures. We demonstrate that intein-mediated protein alternative splicing could be a new strategy to increase protein diversity (that is, functions) without any modification in genetic backgrounds. We also exploited it as a post-translational protein conformation–driven switch of protein functions (for example, as highly specific protein interference).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Paulus, H. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69, 447–496 (2000).
Hirata, R. et al. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H+-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265, 6726–6733 (1990).
Kane, P.M. et al. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science 250, 651–657 (1990).
Perler, F.B. InBase, the Intein Database. Nucleic Acids Res. 30, 383–384 (2002).
Callahan, B.P., Topilina, N.I., Stanger, M.J., Van Roey, P. & Belfort, M. Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat. Struct. Mol. Biol. 18, 630–633 (2011).
Wu, H., Hu, Z. & Liu, X.Q. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 95, 9226–9231 (1998).
Swithers, K.S., Senejani, A.G., Fournier, G.P. & Gogarten, J.P. Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol. Biol. 9, 303 (2009).
Kawasaki, M., Satow, Y., Ohya, Y. & Anraku, Y. Protein splicing in the yeast Vma1 protozyme: evidence for an intramolecular reaction. FEBS Lett. 412, 518–520 (1997).
Southworth, M.W. et al. Control of protein splicing by intein fragment reassembly. EMBO J. 17, 918–926 (1998).
Mootz, H.D. Split inteins as versatile tools for protein semisynthesis. ChemBioChem 10, 2579–2589 (2009).
Perler, F.B. A natural example of protein trans-splicing. Trends Biochem. Sci. 24, 209–211 (1999).
Caspi, J., Amitai, G., Belenkiy, O. & Pietrokovski, S. Distribution of split DnaE inteins in cyanobacteria. Mol. Microbiol. 50, 1569–1577 (2003).
Pietrokovski, S. Intein spread and extinction in evolution. Trends Genet. 17, 465–472 (2001).
Dassa, B., London, N., Stoddard, B.L., Schueler-Furman, O. & Pietrokovski, S. Fractured genes: a novel genomic arrangement involving new split inteins and a new homing endonuclease family. Nucleic Acids Res. 37, 2560–2573 (2009).
Iwai, H., Züger, S., Jin, J. & Tam, P.H. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 580, 1853–1858 (2006).
Ellilä, S., Jurvansuu, J.M. & Iwaï, H. Evaluation and comparison of protein splicing by exogenous inteins with foreign exteins in Escherichia coli. FEBS Lett. 585, 3471–3477 (2011).
Aranko, A.S., Züger, S., Buchinger, E. & Iwaï, H. In vivo and in vitro protein ligation by naturally occurring and engineered split DnaE inteins. PLoS ONE 4, e5185 (2009).
Oeemig, J.S., Aranko, A.S., Djupsjöbacka, J., Heinämäki, K. & Iwaï, H. Solution structure of DnaE intein from Nostoc punctiforme: structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS Lett. 583, 1451–1456 (2009).
Liu, Y. & Eisenberg, D. 3D domain swapping: as domains continue to swap. Protein Sci. 11, 1285–1299 (2002).
Züger, S. & Iwai, H. Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat. Biotechnol. 23, 736–740 (2005).
Heinämäki, K., Oeemig, J.S., Pääkkonen, K., Djupsjöbacka, J. & Iwaï, H. NMR resonance assignment of DnaE intein from Nostoc punctiforme. Biomol. NMR Assign. 3, 41–43 (2009); erratum 7, 115–116 (2013).
Muona, M., Aranko, A.S., Raulinaitis, V. & Iwai, H. Segmental isotopic labeling of multi-domain and fusion proteins by protein trans-splicing in vivo and in vitro. Nat. Protoc. 5, 574–587 (2010).
Klabunde, T., Sharma, S., Telenti, A., Jacobs, W.R. & Sacchettini, J.C. Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol. 5, 31–36 (1998).
Mizutani, R. et al. Protein-splicing reaction via a thiazolidine intermediate: crystal structure of the VMA1-derived endonuclease bearing the N and C-terminal propeptides. J. Mol. Biol. 316, 919–929 (2002).
Sun, P. et al. Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing. J. Mol. Biol. 353, 1093–1105 (2005).
Oeemig, J.S., Zhou, D., Kajander, T., Wlodawer, A. & Iwaï, H. NMR and crystal structures of the Pyrococcus horikoshii RadA intein guide a strategy for engineering a highly efficient and promiscuous intein. J. Mol. Biol. 421, 85–99 (2012).
Ogihara, N.L. et al. Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc. Natl. Acad. Sci. USA 98, 1404–1409 (2001).
Bennett, M.J., Choe, S. & Eisenberg, D. Domain swapping: entangling alliances between proteins. Proc. Natl. Acad. Sci. USA 91, 3127–3131 (1994).
Zettler, J., Schütz, V. & Mootz, H.D. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 583, 909–914 (2009).
Nogami, S. et al. Homing at an extragenic locus mediated by VDE (PI-SceI) in Saccharomyces cerevisiae. Yeast 19, 773–782 (2002).
Tori, K. & Perler, F.B. The Arthrobacter species FB24 Arth_1007 (DnaB) intein is a pseudogene. PLoS ONE 6, e26361 (2011).
Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).
Clerissi, C., Grimsley, N. & Desdevises, Y. Genetic exchanges of inteins between prasinoviruses (phycodnaviridae). Evolution 67, 18–33 (2013).
Dassa, B., Amitai, G., Caspi, J., Schueler-Furman, O. & Pietrokovski, S. Trans protein splicing of cyanobacterial split inteins in endogenous and exogenous combinations. Biochemistry 46, 322–330 (2007).
Appleby-Tagoe, J.H. et al. Highly efficient and more general cis- and trans-splicing inteins through sequential directed evolution. J. Biol. Chem. 286, 34440–34447 (2011).
Gogarten, J.P., Senejani, A.G., Zhaxybayeva, O., Olendzenski, L. & Hilario, E. Inteins: structure, function, and evolution. Annu. Rev. Microbiol. 56, 263–287 (2002).
Clerissi, C., Desdevises, Y. & Grimsley, N. Prasinoviruses of the marine green alga Ostreococcus tauri are mainly species specific. J. Virol. 86, 4611–4619 (2012).
Paulus, H. Inteins as targets for potential antimycobacterial drugs. Front. Biosci. 8, s1157–s1165 (2003).
Liu, X.-Q. & Yang, J. Prp8 intein in fungal pathogens: target for potential antifungal drugs. FEBS Lett. 572, 46–50 (2004).
Amstutz, P., Forrer, P., Zahnd, C. & Plückthun, A. In vitro display technologies: novel developments and applications. Curr. Opin. Biotechnol. 12, 400–405 (2001).
Tori, K. & Perler, F.B. Expanding the definition of class 3 inteins and their proposed phage origin. J. Bacteriol. 193, 2035–2041 (2011).
Aranko, A.S., Oeemig, J.S. & Iwaï, H. Structural basis for protein trans-splicing by a bacterial intein-like domain: protein ligation without nucleophilic residues. FEBS J. 280, 3256–3269 (2013).
Muona, M., Aranko, A.S. & Iwai, H. Segmental isotopic labelling of a multi-domain protein by protein ligation using protein trans-splicing. ChemBioChem 9, 2958–2961 (2008).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Ericsson, U.B., Hallberg, B.M., Detitta, G.T., Dekker, N. & Nordlund, P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 (2006).
Acknowledgements
This work was supported by the Academy of Finland (131413 and 137995) and the Sigrid Jusélius Foundation and Biocenter Finland (to H.I., for crystallization, MS and NMR facilities at the Institute of Biotechnology). A.S.A. and J.S.O. acknowledge Viikki Doctoral Programme in Molecular Biosciences and the National Doctoral Programme in Informational and Structural Biology for financial support, respectively. The authors thank G. Volkmann for his help at the very early stage of the project. The authors also thank F. Perler for providing us with the latest InBase data set and S. Ferkau and C. Albert for their technical support.
Author information
Authors and Affiliations
Contributions
A.S.A. and H.I. designed and performed experiments, analyzed data and prepared the manuscript; T.K., J.S.O. and H.I. performed crystallographic experiments and analyzed diffraction data; H.I. and J.S.O. performed NMR measurements and analyzed NMR data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3, Supplementary Figures 1–9 and Supplementary Note. (PDF 1779 kb)
Rights and permissions
About this article
Cite this article
Aranko, A., Oeemig, J., Kajander, T. et al. Intermolecular domain swapping induces intein-mediated protein alternative splicing. Nat Chem Biol 9, 616–622 (2013). https://doi.org/10.1038/nchembio.1320
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1320
This article is cited by
-
An alternative domain-swapped structure of the Pyrococcus horikoshii PolII mini-intein
Scientific Reports (2021)
-
Small Molecule-Induced Domain Swapping as a Mechanism for Controlling Protein Function and Assembly
Scientific Reports (2017)
-
Domain swapping oligomerization of thermostable c-type cytochrome in E. coli cells
Scientific Reports (2016)
-
Transient misfolding dominates multidomain protein folding
Nature Communications (2015)