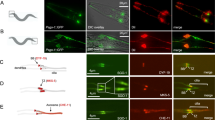
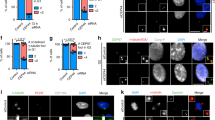
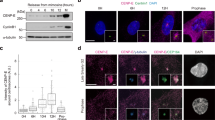
Abstract
Centrins are calmodulin-like proteins that function in the duplication of microtubule-organizing centres. Here we describe a new function of the yeast centrin Cdc31. We show that overproduction of a sequence, termed CID, in the carboxy-terminal domain of the nuclear export factor Sac3 titrates Cdc31, causing a dominant-lethal phenotype and a block in spindle pole body (SPB) duplication. Under normal conditions, the CID motif recruits Cdc31 and Sus1 (a subunit of the SAGA transcription complex) to the Sac3–Thp1 complex, which functions in mRNA export together with specific nucleoporins at the nuclear basket. A previously reported cdc31 temperature-sensitive allele, which is neither defective in SPB duplication nor Kic1 kinase activation, induces mRNA export defects. Thus, Cdc31 has an unexpected link to the mRNA export machinery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Fahrenkrog, B. & Aebi, U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nature Rev. Mol. Cell Biol. 4, 757–766 (2003).
Maniatis, T. & Reed, R. An extensive network of coupling between gene expression machines. Nature 416, 499–506 (2002).
Reed, R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15, 326–331 (2003).
Stutz, F. & Izaurralde, E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 13, 319–327 (2003).
Reed, R. & Hurt, E.C. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108, 523–531 (2002).
Jensen, T.H., Dower, K., Libri, D. & Rosbash, M. Early formation of mRNP. License for export or quality control? Mol. Cell 11, 1129–1138 (2003).
Zenklusen, D., Vinciguerra, P., Strahm, Y. & Stutz, F. The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 21, 4219–4232 (2001).
Sträßer, K. & Hurt, E.C. Yra1p, a conserved nuclear RNA binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19, 410–420 (2000).
Stutz, F. et al. REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6, 638–650 (2000).
Rodrigues, J.P. et al. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA 98, 1030–1035 (2001).
Sträßer, K. et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308 (2002).
Chávez, S. et al. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19, 5824–5834 (2000).
Hurt, E., Luo, M.J., Rother, S., Reed, R. & Strasser, K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl Acad. Sci. USA 101, 1858–1862 (2004).
Kim, M., Ahn, S.H., Krogan, N.J., Greenblatt, J.F. & Buratowski, S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23, 354–364 (2004).
Zenklusen, D., Vinciguerra, P., Wyss, J.C. & Stutz, F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 22, 8241–8253 (2002).
Huertas, P. & Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003).
Fischer, T. et al. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21, 5843–5852 (2002).
Gallardo, M., Luna, R., Erdjument-Bromage, H., Tempst, P. & Aguilera, A. The Nab2p and the Thp1p–Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 278, 24225–24232 (2003).
Lei, P. et al. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol. Biol. Cell 14, 836–847 (2003).
Rodriguez-Navarro, S. et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116, 75–86 (2004).
Novick, P., Osmond, B.C. & Botstein, D. Suppressors of yeast actin mutations. Genetics 121, 659–674 (1989).
Jones, A.L. et al. SAC3 may link nuclear protein export to cell cycle progression. Proc. Natl Acad. Sci. USA 97, 3224–3229 (2000).
Gallardo, M. & Aguilera, A. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics 157, 79–89 (2001).
Biggins, S. & Rose, M.D. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 125, 843–852 (1994).
Spang, A., Courtney, I., Fackler, U., Matzner, M. & Schiebel, E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123, 405–416 (1993).
Spang, A., Courtney, I., Grein, K., Matzner, M. & Schiebel, E. The cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128, 863–878 (1995).
Donaldson, A.D. & Kilmartin, J.V. Spc42p: A phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J. Cell Biol. 132, 887–901 (1996).
Kilmartin, J.V. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162, 1211–1221 (2003).
Rout, M.P. et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651 (2000).
Ivanovska, I. & Rose, M.D. Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics 157, 503–518 (2001).
Belli, G., Gari, E., Piedrafita, L., Aldea, M. & Herrero, E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26, 942–947 (1998).
Baum, P., Furlong, C. & Byers, B. Yeast gene required for spindle pole body duplication: Homology of its product with calcium binding proteins. Proc. Natl Acad. Sci. USA 83, 5512–5516 (1986).
Paoletti, A. et al. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell 14, 2793–2808 (2003).
Araki, M. et al. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276, 18665–18672 (2001).
Pulvermuller, A. et al. Calcium-dependent assembly of centrin-G-protein complex in photoreceptor cells. Mol. Cell. Biol. 22, 2194–2203 (2002).
Sullivan, D.S., Biggins, S. & Rose, M.D. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143, 751–765 (1998).
Siniossoglou, S. et al. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84, 265–275 (1996).
Geier, B.M., Wiech, H. & Schiebel, E. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J. Biol. Chem. 271, 28366–28374 (1996).
Huisinga, K.L. & Pugh, B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13, 573–585 (2004).
Echevarria, W., Leite, M.F., Guerra, M.T., Zipfel, W.R. & Nathanson, M.H. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nature Cell Biol. 5, 440–446 (2003).
Gavin, A.-C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Baßler, J. et al. Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell 8, 517–529 (2001).
Acknowledgements
We would like to thank the following for technical assistance: E.-M. Lammer, M. Schneider and S. Brettschneider in the Hurt laboratory and S. Merker and P. Ihrig in the Lechner laboratory (Mass Spectrometry Unit, BZH, Heidelberg). E.C.H. was supported by grants from the Deutsche Forschungsgemeinschaft (SFB638, Leibniz-Programm) and Fonds der Chemischen Industrie.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Fischer, T., Rodríguez-Navarro, S., Pereira, G. et al. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol 6, 840–848 (2004). https://doi.org/10.1038/ncb1163
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1163
This article is cited by
-
Dynamic modules of the coactivator SAGA in eukaryotic transcription
Experimental & Molecular Medicine (2020)
-
Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway
Nature Communications (2020)
-
Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in Arabidopsis
Nature Plants (2020)
-
mRNA export in the apicomplexan parasite Toxoplasma gondii: emerging divergent components of a crucial pathway
Parasites & Vectors (2018)
-
Nuclear roles for cilia-associated proteins
Cilia (2017)