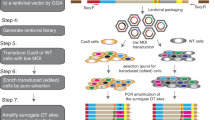
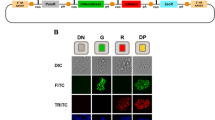
Abstract
Short hairpin RNAs (shRNAs) are versatile tools for analyzing loss-of-function phenotypes in vitro and in vivo1. However, their use for studying genes involved in proliferation and survival, which are potential therapeutic targets in cancer and other diseases, is confounded by the strong selective advantage of cells in which shRNA expression is inefficient. We therefore developed a toolkit that combines Tet-regulated miR30-shRNA technology, robust transactivator expression and two fluorescent reporters to track and isolate cells with potent target knockdown. We demonstrated that this system improves the study of essential genes and was sufficiently robust to eradicate aggressive cancer in mice by suppressing a single gene. Further, we applied this system for in vivo negative-selection screening with pooled shRNAs and propose a streamlined, inexpensive workflow that will facilitate the use of RNA interference (RNAi) for the identification and evaluation of essential therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Martin, S.E. & Caplen, N.J. Applications of RNA interference in mammalian systems. Annu. Rev. Genomics Hum. Genet. 8, 81–108 (2007).
Hannon, G.J. RNA interference. Nature 418, 244–251 (2002).
Brummelkamp, T.R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J. & Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16, 948–958 (2002).
Hemann, M.T. et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33, 396–400 (2003).
Dickins, R.A. et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37, 1289–1295 (2005).
Barbie, D.A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Luo, J. et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 (2009).
Ngo, V.N. et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 (2006).
Scholl, C. et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821–834 (2009).
Stegmeier, F., Hu, G., Rickles, R.J., Hannon, G.J. & Elledge, S.J. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 102, 13212–13217 (2005).
Gossen, M. et al. Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766–1769 (1995).
Lund, A.H., Duch, M. & Pedersen, F.S. Transcriptional silencing of retroviral vectors. J. Biomed. Sci. 3, 365–378 (1996).
Ellis, J., Hotta, A. & Rastegar, M. Retrovirus silencing by an epigenetic TRIM. Cell 131, 13–14 (2007).
Markstein, M., Pitsouli, C., Villalta, C., Celniker, S.E. & Perrimon, N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483 (2008).
Pikaart, M.J., Recillas-Targa, F. & Felsenfeld, G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 12, 2852–2862 (1998).
Agha-Mohammadi, S. et al. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J. Gene Med. 6, 817–828 (2004).
Wold, M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997).
Hochedlinger, K., Yamada, Y., Beard, C. & Jaenisch, R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–477 (2005).
Weinstein, I.B. Cancer. Addiction to oncogenes–the Achilles heal of cancer. Science 297, 63–64 (2002).
Das, A.T. et al. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 279, 18776–18782 (2004).
Meacham, C.E., Ho, E.E., Dubrovsky, E., Gertler, F.B. & Hemann, M.T. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat. Genet. 41, 1133–1137 (2009).
Silva, J.M. et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319, 617–620 (2008).
Schlabach, M.R. et al. Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 (2008).
Bassik, M.C. et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat. Methods 6, 443–445 (2009).
Shin, K.J. et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. USA 103, 13759–13764 (2006).
Zuber, J. et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 23, 877–889 (2009).
Huesken, D. et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 23, 995–1001 (2005).
McCurrach, M.E. & Lowe, S.W. Methods for studying pro- and antiapoptotic genes in nonimmortal cells. Methods Cell Biol. 66, 197–227 (2001).
Schmitt, C.A. et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1, 289–298 (2002).
Taylor, J., Schenck, I., Blankenberg, D. & Nekrutenko, A. Using Galaxy to perform large-scale interactive data analyses. Curr. Protoc. Bioinformatics 10, 10.15 (2007).
Acknowledgements
We thank R. Dickins and C. Miething for discussions in setting up this system, as well as A. Lujambio, A. Rappaport, M. Saborowski and C. Vakoc for testing TRMPV in other models. We also thank Y. Dou (Univ. of Michigan) for providing human MLL-AF9. We gratefully acknowledge B. Ma and S. Muller for excellent technical assistance. We also thank E. Hodges, K. Chang, M. Rooks and the McCombie laboratory for help with Solexa sequencing as well as A. Gordon for bioinformatics support. P. Moody and T. Spencer were of great assistance in flow cytometry. Finally, we thank S. Kogan for histology. This work was supported by the Howard Hughes Medical Institute, the Starr Foundation and the Don Monti Memorial Research Foundation. J.Z. is the Andrew Seligson Memorial Fellow at Cold Spring Harbor Laboratory, and K.M. is the Robert and Teresa Lindsay Fellow of the Watson School of Biological Sciences. L.E.D. is supported by an Overseas Biomedical Research Fellowship of the National Health and Medical Research Council of Australia.
Author information
Authors and Affiliations
Contributions
J.Z. and K.M. designed and performed experiments. C.F. and L.E.D. contributed new reagents and performed experiments. M.J.T. managed mouse monitoring and husbandry. G.J.H. and S.W.L. supervised this project. J.Z., K.M. and S.W.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Table 2 and Supplementary Figs. 1–8 (PDF 3418 kb)
Rights and permissions
About this article
Cite this article
Zuber, J., McJunkin, K., Fellmann, C. et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 29, 79–83 (2011). https://doi.org/10.1038/nbt.1720
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1720
This article is cited by
-
UTX loss alters therapeutic responses in KMT2A-rearranged acute myeloid leukemia
Leukemia (2023)
-
ABCC1 and glutathione metabolism limit the efficacy of BCL-2 inhibitors in acute myeloid leukemia
Nature Communications (2023)
-
Integrated multi-omics analyses reveal homology-directed repair pathway as a unique dependency in near-haploid leukemia
Blood Cancer Journal (2023)
-
Study on the dominant mechanism of direct hole oxidation for the photodegradation of tetracycline
Environmental Science and Pollution Research (2023)
-
YAP induces an oncogenic transcriptional program through TET1-mediated epigenetic remodeling in liver growth and tumorigenesis
Nature Genetics (2022)