Abstract
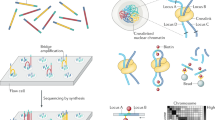
Recent advances in high-throughput DNA sequencing technologies have enabled order-of-magnitude improvements in both cost and throughput. Here we report the use of single-molecule methods to sequence an individual human genome. We aligned billions of 24- to 70-bp reads (32 bp average) to ∼90% of the National Center for Biotechnology Information (NCBI) reference genome, with 28× average coverage. Our results were obtained on one sequencing instrument by a single operator with four data collection runs. Single-molecule sequencing enabled analysis of human genomic information without the need for cloning, amplification or ligation. We determined ∼2.8 million single nucleotide polymorphisms (SNPs) with a false-positive rate of less than 1% as validated by Sanger sequencing and 99.8% concordance with SNP genotyping arrays. We identified 752 regions of copy number variation by analyzing coverage depth alone and validated 27 of these using digital PCR. This milestone should allow widespread application of genome sequencing to many aspects of genetics and human health, including personal genomics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Venter, J.C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007).
Wheeler, D.A. et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 (2008).
Ley, T.J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008).
Wang, J. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Kim, J.I. et al. A highly annotated whole-genome sequence of a Korean individual. Nature advance online publication doi:10.1038/nature08211 (8 July 2009).
Ahn, S.-M. et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. published online, doi:10.1101/gr.092197.109 (26 May 2009).
Braslavsky, I., Hebert, B., Kartalov, E. & Quake, S.R. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci. USA 100, 3960–3964 (2003).
Greenleaf, W.J. & Block, S.M. Single-molecule, motion-based DNA sequencing using RNA polymerase. Science 313, 801 (2006).
Harris, T.D. et al. Single-molecule DNA sequencing of a viral genome. Science 320, 106–109 (2008).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Rumble, S.M. et al. SHRiMP: Accurate Mapping of Short Color-space Reads. PLOS Comput. Biol. 5, 1000386 (2009).
Daines, B. et al. High-throughput multiplex sequencing to discover copy number variants in Drosophila. Genetics published online, doi:10.1534/genetics.109.103218 (15 June 2009).
Herman, D.S. et al. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat. Methods 6, 507–510 (2009).
Xie, C. & Tammi, M.T. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10, 80 (2009).
Iafrate, A.J. et al. Detection of large-scale variation in the human genome. Nat. Genet. 36, 949–951 (2004).
Qin, J., Jones, R.C. & Ramakrishnan, R. Studying copy number variations using a nanofluidic platform. Nucleic Acids Res. 36, e116 (2008).
Vijg, J., Busuttil, R.A., Bahar, R. & Dollé, M.E. Aging and genome maintenance. Ann. NY Acad. Sci. 1055, 35–47 (2005).
Janeway, C. Immunobiology (Garland Science, New York, 2004).
Acknowledgements
We are grateful to V. Natu and J. Coller of the Stanford Functional Genomics Facility for performing the Illumina SNP analysis, R.A. White for assistance with dPCR assays, and A. Sidow for the use of the Covaris sonicator. We acknowledge National Science Foundation award CNS-0619926 for computer resources funding the Bio-X2 cluster, and the National Institutes of Health Pioneer Award (to S.R.Q.).
Author information
Authors and Affiliations
Contributions
N.F.N. prepared the libraries, performed the sequencing and wrote the manuscripts. D.P. developed the data analysis algorithms, performed the computations and wrote the manuscript. S.R.Q. designed the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.P. owns shares of Helicos. S.R.Q. is a founder, shareholder and consultant for Helicos and Fluidigm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 126 kb)
Rights and permissions
About this article
Cite this article
Pushkarev, D., Neff, N. & Quake, S. Single-molecule sequencing of an individual human genome. Nat Biotechnol 27, 847–850 (2009). https://doi.org/10.1038/nbt.1561
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1561
This article is cited by
-
A systematic evaluation of copy number alterations detection methods on real SNP array and deep sequencing data
BMC Bioinformatics (2019)
-
Single-molecule real-time sequencing facilitates the analysis of transcripts and splice isoforms of anthers in Chinese cabbage (Brassica rapa L. ssp. pekinensis)
BMC Plant Biology (2019)
-
Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures
Nature Biotechnology (2018)
-
Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants
Scientific Reports (2018)
-
Highly accurate fluorogenic DNA sequencing with information theory–based error correction
Nature Biotechnology (2017)