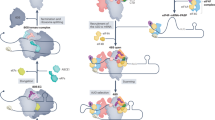
Abstract
Eukaryotic mRNAs contain a 5′ cap structure that is crucial for recruitment of the translation machinery and initiation of protein synthesis. mRNA recognition is thought to require direct interactions between eukaryotic initiation factor 4E (eIF4E) and the mRNA cap. However, translation of numerous capped mRNAs remains robust during cellular stress, early development, and cell cycle progression1 despite inactivation of eIF4E. Here we describe a cap-dependent pathway of translation initiation in human cells that relies on a previously unknown cap-binding activity of eIF3d, a subunit of the 800-kilodalton eIF3 complex. A 1.4 Å crystal structure of the eIF3d cap-binding domain reveals unexpected homology to endonucleases involved in RNA turnover, and allows modelling of cap recognition by eIF3d. eIF3d makes specific contacts with the cap, as exemplified by cap analogue competition, and these interactions are essential for assembly of translation initiation complexes on eIF3-specialized mRNAs2 such as the cell proliferation regulator c-Jun (also known as JUN). The c-Jun mRNA further encodes an inhibitory RNA element that blocks eIF4E recruitment, thus enforcing alternative cap recognition by eIF3d. Our results reveal a mechanism of cap-dependent translation that is independent of eIF4E, and illustrate how modular RNA elements work together to direct specialized forms of translation initiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gingras, A. C., Raught, B. & Sonenberg, N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 (1999)
Lee, A. S., Kranzusch, P. J. & Cate, J. H. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114 (2015)
Sonenberg, N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 86, 178–183 (2008)
Sonenberg, N., Morgan, M. A., Merrick, W. C. & Shatkin, A. J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl Acad. Sci. USA 75, 4843–4847 (1978)
Pause, A. et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371, 762–767 (1994)
Gingras, A. C. et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13, 1422–1437 (1999)
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012)
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012)
Gandin, V. et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 26, 636–648 (2016)
Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010)
Shatkin, A. J. Capping of eucaryotic mRNAs. Cell 9, 645–653 (1976)
Andreev, D. E. et al. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 37, 6135–6147 (2009)
Wolfe, A. L. et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513, 65–70 (2014)
Chang, J. H. et al. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nat. Struct. Mol. Biol. 19, 1011–1017 (2012)
Jiao, X., Chang, J. H., Kilic, T., Tong, L. & Kiledjian, M. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell 50, 104–115 (2013)
Xiang, S. et al. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458, 784–788 (2009)
Sonenberg, N., Morgan, M. A., Testa, D., Colonno, R. J. & Shatkin, A. J. Interaction of a limited set of proteins with different mRNAs and protection of 5′-caps against pyrophosphatase digestion in initiation complexes. Nucleic Acids Res. 7, 15–29 (1979)
Lindqvist, L., Imataka, H. & Pelletier, J. Cap-dependent eukaryotic initiation factor–m RNA interactions probed by cross-linking. RNA 14, 960–969 (2008)
Locker, N., Easton, L. E. & Lukavsky, P. J. Affinity purification of eukaryotic 48S initiation complexes. RNA 12, 683–690 (2006)
Zhang, L., Pan, X. & Hershey, J. W. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J. Biol. Chem. 282, 5790–5800 (2007)
Blau, L. et al. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc. Natl Acad. Sci. USA 109, E2875–E2884 (2012)
Hay, N. & Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 (2004)
Xue, S. et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 (2015)
Wisdom, R., Johnson, R. S. & Moore, C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18, 188–197 (1999)
des Georges, A. et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525, 491–495 (2015)
Sun, C. et al. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl Acad. Sci. USA 108, 20473–20478 (2011)
Lee, A. S., Burdeinick-Kerr, R. & Whelan, S. P. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. USA 110, 324–329 (2013)
Kranzusch, P. J. et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 158, 1011–1021 (2014)
Kranzusch, P. J. et al. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Terwilliger, T. C. Reciprocal-space solvent flattening. Acta Crystallogr. D 55, 1863–1871 (1999)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Acknowledgements
The authors thank J. Berger and K. Chat for discussions. X-ray data were collected at Beamline 8.3.1 of the Lawrence Berkeley National Laboratory Advanced Light Source (ALS), supported in part by the UC Office of the President, Multicampus Research Programs and Initiatives grant MR-15-328599 and the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation. The authors are grateful to J. Holton, G. Meigs (ALS), and T. Doukov (SSRL) for help with S-SAD data collection. This work used the Vincent J. Proteomics/Mass Spectrometry Laboratory at UC Berkeley, supported in part by NIH S10 Instrumentation Grant S10RR025622. This work was funded by the NIGMS Center for RNA Systems Biology (P50-GM201706). J.A.D. is an HHMI Investigator. A.S.Y.L. is supported as an American Cancer Society Postdoctoral Fellow (PF-14-108-01-RMC) and P.J.K. is supported as an HHMI Fellow of the Life Sciences Research Foundation.
Author information
Authors and Affiliations
Contributions
Project and experiments were conceived and designed by A.S.Y.L. with consultation with J.H.D.C. Protein purification, biochemistry, and cell biology experiments and analyses were performed by A.S.Y.L. Crystallography and structure determination was performed by P.J.K. The manuscript was written by A.S.Y.L., P.J.K. and J.H.D.C, with editing by J.A.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks I. Topisirovic and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 c-Jun expression is unaffected by 4E-BP1 activation.
Representative western blot of 293T cells after 24 h treatment with mTOR inhibitor INK128. The results are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2 Mapping of a C-terminal region of eIF3d that protects the c-Jun 5′ cap structure.
a, Validation of eIF3d subunit identification. eIF3d-cap crosslinking was validated by immunoprecipitation of eIF3d after crosslinking and denaturing the eIF3 complex by boiling in SDS. The result is representative of biological replicates. b, Limited proteolysis of eIF3 crosslinked to 32P-cap-labelled c-Jun 5′ UTR RNA. Full-length and proteolysis fragments of eIF3d are indicated by black and maroon arrows, respectively, on the phosphorimage and Coomassie-stained SDS gels. c, Mass spectrometry identification of trypsinized peptides from limited proteolysis of cap-crosslinked eIF3d. Identified peptides are highlighted in blue. The results in b and c are representative of three independent experiments.
Extended Data Figure 3 Purification of eIF3d cap-binding domain.
a, Alignment is coloured by phylogenetic conservation of amino acid physiochemical property similarity and a cartoon schematic of the eIF3d secondary structure is depicted below the sequences. Colouring begins at 30% conservation (lightest blue). b, Coomassie-blue-stained SDS gel of recombinant N. vitripennis eIF3d S172–F537 protein expressed in E. coli.
Extended Data Figure 4 Structure-based alignment of eIF3d cap-binding domain and DXO cap-endonuclease domain sequences.
Structure-based alignment of eIF3d and DXO sequences according to superposition of eIF3d and DXO (PDB 4J7L) structures15. Alignment is coloured by phylogenetic conservation as in Extended Data Fig. 2, and cartoon schematics of the secondary structures are depicted below the sequences. eIF3d is coloured in blue and magenta as in Fig. 2, and DXO is coloured in green and magenta.
Extended Data Figure 5 Structural details of eIF3d ‘RNA gate’ stabilizing interactions.
a–c, Structural overview of eIF3d with cut-away sections highlighting charged interactions stabilizing the closed ‘RNA gate’ conformation. No significant van der Waals interactions stabilize the closed gate conformation, supporting likely repositioning of the RNA gate before 5′ mRNA cap recognition. Charged interactions occur in three areas: a, at the beginning of the gate insertion sequence (gate beginning); b, at the tip of the unstructured loop (gate tip); and c, at an ‘arginine anchor’ point stabilizing the return of the loop insertion sequence to the α-helix shared with DXO family endonucleases. Residues are numbered according to the human eIF3d sequence, and all positions are conserved between human and N. vitripennis except S292N. eIF3d RNA gate residues are displayed with blue side chains and the residues making stabilizing contacts are coloured in green. 2Fo − Fc map regions are shown at 1.5σ.
Extended Data Figure 6 Packing interactions observed in alternative eIF3d crystal forms.
Cartoon representation of crystallographic packing in eIF3d crystal form 1, 2 and 3 (a, b and c). Crystal forms 1 and 3 have two copies of eIF3d in the asymmetric unit coloured in blue/magenta and green/magenta, respectively; crystal form 2 has only one copy of eIF3d per asymmetric unit. Symmetry-related molecules are depicted in grey. Cut-away zoom illustrates position of the eIF3d RNA gate (red) relative to the nearest symmetry-related molecule. In crystal form 1, the RNA gate is packed against a neighbouring symmetry molecule, but in crystal forms 2 and 3, the RNA gate is positioned towards a major solvent channel. Relative conformation of the RNA gate remains unchanged in either eIF3d crystal form.
Extended Data Figure 7 eIF3d cap-binding activity requires the eIF3-recruitment stem–loop RNA.
Phosphorimage of SDS gel resolving RNase-protected 32P-cap-labelled c-Jun stem–loop RNA crosslinked to eIF3 subunits. The result is representative of three independent experiments.
Extended Data Figure 8 Incorporation of HA epitope-tagged eIF3d into translation initiation complexes.
a, Coomassie-blue-stained SDS gel of recombinant eIF3 containing wild-type or helix α5- or α11-mutated eIF3d. b, Representative native agarose gel electrophoresis of recombinant wild-type and mutant eIF3 complexes bound to the c-Jun stem–loop. c, Polysome profiles of untransfected 293T cells, plotted as relative absorbance at 254 nm versus elution fractions. d, Western blot analysis of eIF3d and the small (rpS19) and large (rpLP0) ribosomal subunits. The results in b–d are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 9 eIF4E recognizes the 5′ end of the c-Jun mRNA less efficiently than ACTB mRNA.
a, Coomassie-blue-stained SDS gel of recombinant human eIF4E expressed in E. coli. b, Phosphorimage of SDS gel resolving RNase-protected 32P-cap-labelled ACTB or c-Jun 5′ UTR RNA crosslinked to eIF4E. The result is representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 and Supplementary Table 1. (PDF 647 kb)
Rights and permissions
About this article
Cite this article
Lee, A., Kranzusch, P., Doudna, J. et al. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99 (2016). https://doi.org/10.1038/nature18954
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18954
This article is cited by
-
FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells
Genome Biology (2024)
-
Akt enhances the vulnerability of cancer cells to VCP/p97 inhibition-mediated paraptosis
Cell Death & Disease (2024)
-
The molecular basis of translation initiation and its regulation in eukaryotes
Nature Reviews Molecular Cell Biology (2024)
-
System-wide analysis of RNA and protein subcellular localization dynamics
Nature Methods (2024)
-
N6-Methyladenosine Methylation of mRNA in Cell Apoptosis
Molecular Neurobiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.