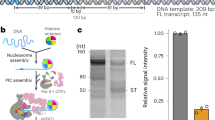
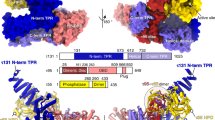
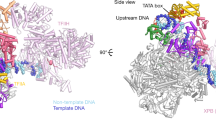
Abstract
The general transcription factor IID (TFIID) plays a central role in the initiation of RNA polymerase II (Pol II)-dependent transcription by nucleating pre-initiation complex (PIC) assembly at the core promoter. TFIID comprises the TATA-binding protein (TBP) and 13 TBP-associated factors (TAF1–13), which specifically interact with a variety of core promoter DNA sequences. Here we present the structure of human TFIID in complex with TFIIA and core promoter DNA, determined by single-particle cryo-electron microscopy at sub-nanometre resolution. All core promoter elements are contacted by subunits of TFIID, with TAF1 and TAF2 mediating major interactions with the downstream promoter. TFIIA bridges the TBP–TATA complex with lobe B of TFIID. We also present the cryo-electron microscopy reconstruction of a fully assembled human TAF-less PIC. Superposition of common elements between the two structures provides novel insights into the general role of TFIID in promoter recognition, PIC assembly, and transcription initiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under codes EMD-3304 (TFIID-IIA–SCP complex), EMD-3305 (locally refined BC-core of TFIID-IIA–SCP complex), EMD-3306 (locally refined lobe C of TFIID-IIA–SCP complex), and EMD-3307 (TAF-less PIC). Model coordinates have been deposited in the Protein Data Bank (PDB) under accession number 5FUR.
Change history
20 April 2016
A Correction to this paper has been published: https://doi.org/10.1038/nature17984
References
Levine, M., Cattoglio, C. & Tjian, R. Looping back to leap forward: transcription enters a new era. Cell 157, 13–25 (2014)
Thomas, M. C. & Chiang, C. M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 (2006)
Buratowski, S., Hahn, S., Guarente, L. & Sharp, P. A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–561 (1989)
Burley, S. K. & Roeder, R. G. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799 (1996)
Albright, S. R. & Tjian, R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242, 1–13 (2000)
Verrijzer, C. P., Chen, J. L., Yokomori, K. & Tjian, R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81, 1115–1125 (1995)
Burke, T. W. & Kadonaga, J. T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11, 3020–3031 (1997)
Chalkley, G. E. & Verrijzer, C. P. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 18, 4835–4845 (1999)
Lee, D. H. et al. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol. Cell. Biol. 25, 9674–9686 (2005)
Theisen, J. W., Lim, C. Y. & Kadonaga, J. T. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol. Cell. Biol. 30, 3471–3479 (2010)
Hisatake, K. et al. Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and interactions of TAFII80 with other TAFs and with general transcription factors. Proc. Natl Acad. Sci. USA 92, 8195–8199 (1995)
Ruppert, S. & Tjian, R. Human TAFII250 interacts with RAP74: implications for RNA polymerase II initiation. Genes Dev. 9, 2747–2755 (1995)
Dubrovskaya, V. et al. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIFβ (RAP30) and incorporation into the TFIID complex. EMBO J. 15, 3702–3712 (1996)
Wu, S. Y. & Chiang, C. M. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276, 34235–34243 (2001)
Xie, X. et al. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380, 316–322 (1996)
Birck, C. et al. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94, 239–249 (1998)
Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000)
Werten, S. et al. Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAF(II)135) and hTAF12 (hTAF(II)20). J. Biol. Chem. 277, 45502–45509 (2002)
Bleichenbacher, M., Tan, S. & Richmond, T. J. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J. Mol. Biol. 332, 783–793 (2003)
Bhattacharya, S., Takada, S. & Jacobson, R. H. Structural analysis and dimerization potential of the human TAF5 subunit of TFIID. Proc. Natl Acad. Sci. USA 104, 1189–1194 (2007)
Wang, X. et al. Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc. Natl Acad. Sci. USA 104, 7839–7844 (2007)
Scheer, E., Delbac, F., Tora, L., Moras, D. & Romier, C. TFIID TAF6-TAF9 complex formation involves the HEAT repeat-containing C-terminal domain of TAF6 and is modulated by TAF5 protein. J. Biol. Chem. 287, 27580–27592 (2012)
Anandapadamanaban, M. et al. High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nature Struct. Mol. Biol . 20, 1008–1014 (2013)
Wang, H., Curran, E. C., Hinds, T. R., Wang, E. H. & Zheng, N. Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module. Cell Res. 24, 1433–1444 (2014)
Trowitzsch, S. et al. Cytoplasmic TAF2–TAF8–TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nature Commun . 6, 6011 (2015)
Andel, F., III, Ladurner, A. G., Inouye, C., Tjian, R. & Nogales, E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286, 2153–2156 (1999)
Brand, M., Leurent, C., Mallouh, V., Tora, L. & Schultz, P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286, 2151–2153 (1999)
Leurent, C. et al. Mapping histone fold TAFs within yeast TFIID. EMBO J. 21, 3424–3433 (2002)
Grob, P. et al. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure 14, 511–520 (2006)
Liu, W. L. et al. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol. Cell 29, 81–91 (2008)
Papai, G. et al. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465, 956–960 (2010)
Bieniossek, C. et al. The architecture of human general transcription factor TFIID core complex. Nature 493, 699–702 (2013)
Cianfrocco, M. A. et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 152, 120–131 (2013)
Juven-Gershon, T., Cheng, S. & Kadonaga, J. T. Rational design of a super core promoter that enhances gene expression. Nature Methods 3, 917–922 (2006)
Kraemer, S. M., Ranallo, R. T., Ogg, R. C. & Stargell, L. A. TFIIA interacts with TFIID via association with TATA-binding protein and TAF40. Mol. Cell. Biol. 21, 1737–1746 (2001)
Gajiwala, K. S. & Burley, S. K. Winged helix proteins. Curr. Opin. Struct. Biol. 10, 110–116 (2000)
Kokubo, T., Swanson, M. J., Nishikawa, J. I., Hinnebusch, A. G. & Nakatani, Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18, 1003–1012 (1998)
Malkowska, M., Kokoszynska, K., Rychlewski, L. & Wyrwicz, L. Structural bioinformatics of the general transcription factor TFIID. Biochimie 95, 680–691 (2013)
Kochan, G. et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl Acad. Sci. USA 108, 7745–7750 (2011)
Bell, B., Scheer, E. & Tora, L. Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8, 591–600 (2001)
Wright, K. J., Marr, M. T., II & Tjian, R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl Acad. Sci. USA 103, 12347–12352 (2006)
He, Y., Fang, J., Taatjes, D. J. & Nogales, E. Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013)
Yakovchuk, P., Gilman, B., Goodrich, J. A. & Kugel, J. F. RNA polymerase II and TAFs undergo a slow isomerization after the polymerase is recruited to promoter-bound TFIID. J. Mol. Biol. 397, 57–68 (2010)
Zhang, Z. et al. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. eLife 4, e07777 (2015)
Gegonne, A., Devaiah, B. N. & Singer, D. S. TAF7: traffic controller in transcription initiation. Transcription 4, 29–33 (2013)
Plaschka, C. et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 (2015)
Tsai, K. L. et al. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 (2014)
Kim, T. H. et al. A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005)
Carninci, P. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genet. 38, 626–635 (2006)
Cooper, S. J., Trinklein, N. D., Anton, E. D., Nguyen, L. & Myers, R. M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 16, 1–10 (2006)
Pal, M., Ponticelli, A. S. & Luse, D. S. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell 19, 101–110 (2005)
Revyakin, A. et al. Transcription initiation by human RNA polymerase II visualized at single-molecule resolution. Genes Dev. 26, 1691–1702 (2012)
Kim, B. et al. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl Acad. Sci. USA 104, 16068–16073 (2007)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009)
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009)
Scheres, S. H. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3, e03665 (2014)
Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012)
Heymann, J. B. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133, 156–169 (2001)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Garzón, J. I., Kovacs, J., Abagyan, R. & Chacón, P. ADP_EM: fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics 23, 427–433 (2007)
Wriggers, W. Using Situs for the integration of multi-resolution structures. Biophys. Rev . 2, 21–27 (2010)
Lopéz-Blanco, J. R. & Chacón, P. iMODFIT: efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J. Struct. Biol. 184, 261–270 (2013)
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014)
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nature Methods 12, 7–8 (2015)
van Dijk, M. & Bonvin, A. M. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 37, W235–W239 (2009)
Chys, P. & Chacón, P. Random coordinate descent with spinor-matrices and geometric filters for efficient loop closure. J. Chem. Theory Comput. 9, 1821–1829 (2013)
Ramachandran, S., Kota, P., Ding, F. & Dokholyan, N. V. Automated minimization of steric clashes in protein structures. Proteins 79, 261–270 (2011)
Chaudhury, S., Lyskov, S. & Gray, J. J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010)
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 (2013)
Acknowledgements
We thank P. Grob, S. Howes, and R. Zhang for electron microscopy support; T. Houweling for computer support; S. Scheres for technical advice on image processing; A. Patel for discussion; C. Inouye for providing us with recombinant TFIIF; S. Zheng for providing TAF4 mAb; and D. King for providing peptides. We are thankful to R. Tjian and J. Kadonaga for their comments on the manuscript. Computational resources were provided in part by the National Energy Research Scientific Computing Center (DE-AC02-05CH11231). This work was funded by NIGMS (GM63072 to E.N.) and Spanish Ministry of Economy and Competitiveness (BFU2013-44306P to P.C.). R.K.L. was supported by the NIGMS Molecular Biophysics Training Grant (GM008295). E.N. is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
R.K.L. and Y.H. designed and performed the experiments; J.R.L.-B. and P.C. performed structural modelling; J.F. purified the TFIID, Pol II, TFIIE, and TFIIH; R.L., Y.H., and E.N. analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Cryo-EM of the TFIID-IIA–SCP complex.
a, Representative micrograph of frozen-hydrated TFIID-IIA–SCP complexes. Examples of particle picks are indicated by the green circles; 203,163 such picks were made from 1,253 total micrographs. b, Initial classification and refinement scheme for the TFIID-IIA–SCP structure (see Methods). c, Idealized dose-dependent B-factor plot based on cryo-EM data collected on microtubules under similar imaging conditions. This plot was used for the particle polishing step in b. d, e, Fourier shell correlation plot (d) and local resolution estimation (e) for the final reconstruction shown in b.
Extended Data Figure 2 Focused classification and refinement of the promoter-bound BC-core and lobe C of TFIID.
a, b, Scheme for focused classification and refinement of the BC-core region (a) or lobe C region of the TFIID-IIA–SCP structure (b) (see Methods). c, d, Fourier shell correlation plots (c) and local resolution estimations (d) of the BC-core and lobe C maps, corresponding to the final structures shown in a and b, respectively. e, Two-dimensional projections of the refined maps for the full TFIID-IIA–SCP structure (left), locally refined BC-core map (middle), and locally refined lobe C map (right). The maps used to calculate the projections are the same as the final structures in a, b, and Extended Data Fig. 1b, except that all have been low-pass filtered to 10 Å before calculating projections. f, Three-dimensional classification of 56,457 particles into two classes (solid blue and transparent green), following focused alignment to the lobe C region of the structure. The resulting classes have been superposed through their lobe C densities to illustrate the flexibility of lobe B and the upstream region of promoter DNA relative to lobe C and the downstream promoter region. The magnitude of motion within lobe A1 (20 Å) is indicated.
Extended Data Figure 3 Modelling of TBP, TFIIA, and promoter DNA into the cryo-EM density.
a, Previously published reconstructions of TFIID-IIA–SCP in the rearranged state (left; EMDB code 2282) and of free TFIID in the canonical state (right; EMDB code 2287)33. For the former, the densities for TFIIA (orange) and TBP (red) are assigned on the basis of the superposition with the TFIID-IIA–SCP structure from our present study. b, Close-up view of the TBP–TFIIA–TATA module density and fitted structures. The termini of the TBP structure and the three subunits (α, β, and γ) within the TFIIA structure are indicated with circles. In the cell, the α- and β-subunits of TFIIA are translated as a single polypeptide and then are post-translationally cleaved. The location of the long stretch of residues spanning the region between the structured parts of TFIIAα and TFIIAβ (TFIIAαβ 52–329) is indicated as a dashed line. Note that only 34 of the residues within this flexible loop (52–58 and 303–329) are included in the TFIIA construct used for this study. Mutational analysis in yeast has shown that mutation of an isoleucine residue (I23 in humans, I27 in yeast; represented in green spheres) to lysine at the tip of the TFIIA four-helix bundle disrupts the interaction between TFIID and TFIIA35. c, Mapping of the MPE.Fe(II) cleavage pattern for SCP DNA bound to TFIID-IIA, on the basis of data published in ref. 33. d, Mapping of the downstream core element (DCE)9 sequence onto the SCP DNA within the TFIID-IIA–SCP structure from our present study.
Extended Data Figure 4 Structural modelling and conservation of the TAF1 promoter-binding domains.
a, TAF1 WH domain (grey) in complex with promoter DNA (cyan) superposed with the DNA-binding WH domain of the transcription factor E2F4 (PDB accession number 1CF7, magenta) in complex with its cognate DNA, with the alignment based on the protein (left) or DNA (right) components. b, Sequence alignment and secondary structure map of the TAF1 WH domain, used to calculate the conservation scores depicted in Fig. 2c (Hs, Homo sapiens; Dr, Danio rerio; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae). The conserved positively charged residues that are in close proximity to the promoter DNA within the docked structure (K818, R864, K865, K868, and R875) are highlighted in pink. Numbering is based on the human sequence. c, Sequence alignment of a region of the TAF1 DUF3591 corresponding to the internal segment that is missing from the crystal structure and neighbouring residues. The putative Inr-binding domain (1009–1061) within this segment is highlighted in blue. Numbering is based on the human sequence, and abbreviations are the same as in a. d, Three-dimensional structure prediction for the putative TAF1 Inr-binding domain output by the I-TASSER server67. On the left, the residues are coloured in rainbow from N to C termini, with the terminal residues indicated. On the right, the modelling confidence is depicted in terms of the ResQ score (ribbon colour) and B-factor estimation (ribbon thickness) output by I-TASSER67, with high confidence regions represented by thinner blue ribbon and low-confidence regions represented with thicker red ribbon. e, Secondary structure prediction for the sequence modelled in d (H, helix; C, coil).
Extended Data Figure 5 Structural modelling and conservation of TAF2 APD.
a, Structural arrangement of domains (D1–D4) within the TAF2 APD (bottom) compared with that of human ERAP1 (top, PDB accession number 2YD0)39, a member of the M1 family of aminopeptidases to which TAF2 shares homology. b, Domain arrangement of TAF2, including the four subdomains of the APD (D1–D4), and the C-terminal intrinsically disordered region (IDR). c, Rigid-body docking of the best-conserved domains (D1 and D2) of the homologous human ERAP1 confirms the identity of this density. d, Segmented densities and fitted structures for the four subdomains (D1–D4) of the TAF2 APD. e, Sequence alignment and secondary structure map for the putative DNA-binding regions within domain 3 of the TAF2 APD (species abbreviations are the same as in Extended Data Fig. 4b). Conserved residues that are in close proximity to the DNA within the docked structure are highlighted in pink. The stretch that is depicted as a dashed line shares low sequence similarity with known M1 aminopeptidases. Numbering is based on the human sequence.
Extended Data Figure 6 Structural modelling and conservation of TAF6 and putative TAF8 density.
a, Cryo-EM density of the TAF6 dimer with fitted homology models. Putative regions involved in the homodimer interface are labelled. b, Organization of α-helices within the human TAF6 HEAT-like repeat and unaccounted density (green) around the TAF6 homodimer. c, Sequence alignment and secondary structure map of the TAF6 HEAT repeat domain (species abbreviations are the same as in Extended Data Fig. 4b, except that Al is A. locustae). The green region indicates the region that is unmodelled in our structure, with the two predicted C-terminal helices outlined with dashes. Numbering is based on the human sequence. d, Unaccounted density indicative of two α-helices, located between domain 4 of the TAF2 APD and one copy of the TAF6 HEAT domain, which we attribute to TAF8. e, Sequence alignment of a putative TAF2-interaction domain within TAF8 (species abbreviations are the same as in Extended Data Fig. 4b). The last helix of the structurally determined histone fold domain of TAF8 is depicted in dark blue, while the 26 residue stretch that is predicted to be α-helical is shown in light blue with dashed outline. Secondary structure prediction was performed with PSI-PRED71.
Extended Data Figure 7 Modelling of the TFIID-based PIC.
a, TFIID-based PIC model from Fig. 4, with the density for lobe A2 density (yellow) low-pass filtered to 16 Å and displayed at two different intensity thresholds (lower threshold in transparency). Both thresholds are lower than that used to display the density for the promoter-bound BC-core of TFIID. b, Close-up view of putative interactions between RPB1, -2, and -5 of Pol II and TAF1 of TFIID. c, Comparison of the paths of the promoter DNA within the TFIID-IIA–SCP and TAF-less PIC structures. The promoter DNA from the TFIID-IIA–SCP structure is coloured as in Fig. 1, and the promoter DNA from the TAF-less PIC is coloured in green. View is from the top of the model, relative to a. d, Docking of the core mediator coactivator complex (cMed, EMDB code 2786)46, including the mediator head and middle modules, onto the TFIID-based PIC, on the basis of the structure of a cMED-bound initial transcribing complex. e, Docking of the free yeast mediator complex (brown transparency, EMDB code 2634)47 on the basis of alignment with the core mediator shown in c. Lobe A2 of TFIID (yellow) is depicted similarly as in a.
Supplementary information
Structure of promoter-bound TFIID and insight into human PIC assembly
Cryo-EM structure of human TFIID-IIA-SCP and fitting of atomic models. Superposition of common elements between TFIID-IIA-SCP and the TAF-less PIC leads to a model of a TFIID-based PIC. (MOV 29244 kb)
Rights and permissions
About this article
Cite this article
Louder, R., He, Y., López-Blanco, J. et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 (2016). https://doi.org/10.1038/nature17394
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17394
This article is cited by
-
Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
Nature Reviews Genetics (2023)
-
Hierarchical TAF1-dependent co-translational assembly of the basal transcription factor TFIID
Nature Structural & Molecular Biology (2023)
-
Ino2, activator of yeast phospholipid biosynthetic genes, interacts with basal transcription factors TFIIA and Bdf1
Current Genetics (2023)
-
Loss of TAF8 causes TFIID dysfunction and p53-mediated apoptotic neuronal cell death
Cell Death & Differentiation (2022)
-
Taf2 mediates DNA binding of Taf14
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.