Abstract
Knowledge about the underlying genetic architecture of phenotypic traits is needed to understand and predict evolutionary dynamics. The number of causal loci, magnitude of the effects and location in the genome are, however, still largely unknown. Here, we use genome-wide single-nucleotide polymorphism (SNP) data from two large-scale data sets on house sparrows and collared flycatchers to examine the genetic architecture of different morphological traits (tarsus length, wing length, body mass, bill depth, bill length, total and visible badge size and white wing patches). Genomic heritabilities were estimated using relatedness calculated from SNPs. The proportion of variance captured by the SNPs (SNP-based heritability) was lower in house sparrows compared with collared flycatchers, as expected given marker density (6348 SNPs in house sparrows versus 38 689 SNPs in collared flycatchers). Indeed, after downsampling to similar SNP density and sample size, this estimate was no longer markedly different between species. Chromosome-partitioning analyses demonstrated that the proportion of variance explained by each chromosome was significantly positively related to the chromosome size for some traits and, generally, that larger chromosomes tended to explain proportionally more variation than smaller chromosomes. Finally, we found two genome-wide significant associations with very small-effect sizes. One SNP on chromosome 20 was associated with bill length in house sparrows and explained 1.2% of phenotypic variation (VP), and one SNP on chromosome 4 was associated with tarsus length in collared flycatchers (3% of VP). Although we cannot exclude the possibility of undetected large-effect loci, our results indicate a polygenic basis for morphological traits.
Similar content being viewed by others
Introduction
Information about the genetic architecture of phenotypic traits is fundamental to our understanding of how these traits evolve. By revealing the number and effect size of the loci controlling heritable traits, we can improve predictions about trait evolution in natural populations (Barton and Keightley, 2002) and better understand the potential of populations to adapt to environmental change. For example, the HMGA2 gene in Galápagos finches explains a substantial portion of variation in beak morphology, and it was associated with marked character displacement during a severe, acute drought (Lamichhaney et al., 2016). Although there has been an increasing number of studies aiming at identifying genes underlying phenotypic variation in natural populations (reviewed in Slate et al., 2010; Schielzeth and Husby, 2014), the genetic architecture (that is, the number of genes, their effect sizes and location in the genome) of most morphological traits still remains unknown.
A first step in understanding the genetic architecture is to establish whether the trait is heritable, something that traditionally has been done using quantitative genetic methods such as parent–offspring regressions, sib analyses or the ‘animal model’ (Lynch and Walsh, 1998). These models use the expected genetic relatedness among individuals to estimate heritability. However, advances in high-density genotyping have made it possible to use genome-wide marker data to estimate realized genetic relatedness between individuals and, therefore, the ‘genomic heritability’ (Aulchenko et al., 2007; Yang et al., 2010; Zaitlen et al., 2013; Rönnegård et al., 2016). Genome-wide marker data from a large number of individuals can also be used to estimate the proportion of variation in the trait that is tagged by single-nucleotide polymorphism (SNP) arrays, the so-called SNP-based heritability (Yang et al., 2010, 2011b). All these approaches have limitations. For example, pedigree-based heritability estimates (h2ped) require information from known relatives, and heritability values may be biased because of shared environmental factors among relatives. At the same time, SNP-based estimates have been less successful in capturing the full extent of known trait genetic variance. As a result, there is often a gap between heritability estimated from pedigree approaches and heritability estimated obtained by considering significant SNPs from genome-wide association studies (GWAS), which is referred to as the ‘missing heritability’ (Manolio et al., 2009).
The ‘missing heritability’ phenomenon is partly a result of very stringent criteria for determining that a SNP contributes significantly to the trait variance. All SNPs considered jointly explain a much higher proportion of the variance than individually significant SNPs considered jointly (Yang et al., 2010). However, this method requires only unrelated individuals to be used, substantially reducing sample size. To alleviate this, Zaitlen et al. (2013) developed a method to estimate both the proportion of trait variance explained by genotyped SNPs (SNP-based heritability—h2g) and the ‘total narrow-sense heritability’ (h2gkin), which is equivalent to the traditional pedigree-based heritability (Zaitlen et al., 2013).
Given high enough marker density, kinship coefficients can also be estimated on a more regional scale instead of a genome-wide scale. For example, Yang et al. (2011a) proposed partitioning genetic variance of traits onto chromosomes. This method can provide novel insights into the genetic architecture of traits because it is expected that, under a polygenic model, chromosome size should scale positively with the amount of genetic variation explained by that chromosome. Chromosomes that contribute a disproportionate amount of variation, given their size, can therefore indicate the presence of large-effect loci on that chromosome or, alternatively, a cluster of loci of small effect (Schielzeth and Husby, 2014).
Ultimately, we are interested in understanding how evolutionary forces act on complex traits. Genome-wide association methods have been extensively used in human and livestock studies to detect causal loci (for example, Goddard and Hayes, 2009; Yang et al., 2010), and the decreasing cost of genotyping many individuals at thousands of loci means that GWAS are increasingly applied in studies of nonmodel organisms (for example, Johnston et al., 2014; Barson et al., 2015; Husby et al., 2015; Santure et al., 2015). Some of these studies have been successful in identifying large-effect loci (Johnston et al., 2014; Barson et al., 2015), whereas others have failed to identify genome-wide significant variants (Santure et al., 2013). Even in cases where significant variants have been detected, they only explain a relatively small proportion of the phenotypic variance (for example, Bérénos et al., 2015; Husby et al., 2015).
Traditionally, GWAS do not utilize repeated measurements of the same individuals, but many long-term ecological studies follow individuals throughout their lifetime and re-measure phenotypic traits over ontogeny. This adds additional information that could be used in GWAS, and Rönnegård et al. (2016) recently developed a method to incorporate such repeated measures in a GWAS framework. Adding repeated measures can lead to increased power if there are large annual variations in the expression of the trait or unbalanced records per individual. As many GWAS of natural populations suffer from a lack of power as a result of low sample size (for example, Kardos et al., 2016), incorporating repeated measures can therefore be a useful way to increase power to detect QTLs (Rönnegård et al., 2016).
In this study, we take advantage of genomic resources that have recently become available for house sparrows (Passer domesticus; Hagen et al., 2013) and collared flycatchers (Ficedula albicollis; Ellegren et al., 2012), two well-studied model passerine species in evolutionary biology and ecology (Anderson, 2006; Qvarnström et al., 2010). Of relevance to the present study, Hagen et al. (2013) designed a custom Illumina (San Diego, CA, USA) 10K SNP array for house sparrows and Kawakami et al. (2014) designed a custom Illumina 50K SNP array for collared flycatchers. These arrays have an average marker density of one SNP per 100 000 bp for house sparrows (Hagen et al., 2013) and one SNP per 22 000 bp for collared flycatchers. These genomic resources, together with the phenotypic data collected, offer the opportunity to examine the genetic architecture of phenotypic traits.
House sparrows and collared flycatchers group into different phylogenetic clades within Passeriformes, Passeridae and Muscicapidae that diverged ~50 million years ago (Jarvis et al., 2014). Comparing the genetic architecture of different phenotypic traits in these two species gives the opportunity to identify patterns of genetic architecture of phenotypic traits within passerines. Here, we first aimed at estimating genomic heritabilities of morphological traits using both genome-wide and chromosome-specific approaches (see Table 1). Second, we used a recently developed method (Zaitlen et al., 2013) to estimate the proportion of genetic variance captured by the SNP arrays. To identify SNPs associated with the traits studied, we carried out GWAS using both repeated phenotypic measures and mean phenotypic values. Finally, we examined whether the genetic architecture is concordant across similar traits in the two species and across different approaches.
Materials and methods
Study populations and phenotypic data
Phenotypic data from house sparrows were collected as part of a long-term individual-based study on four islands in northern Norway—Aldra (66°25′N, 13°04′E), Hestmannøy (66°33′N, 12°50′E), Leka (65°06′N, 11°38′E) and Vega (65°40′N, 11°55′E)—that has been running since 1993 (for example, Jensen et al., 2008). Five phenotypic characters were measured in adults of both sexes (Figure 1): tarsus length, wing length, body mass, bill depth and bill length. In addition, both total badge size and visible badge size (see Figure 1) were measured in adult males as there is evidence of different mechanisms for the expression of these two traits, and they may act as different signals (Veiga, 1996). Total badge size was measured as the square root of the area covered by black feathers and feathers with black bases and gray tips on the throat and the chest, whereas visible badge size was measured as the square root of the area covered by completely black feathers, that is, excluding the feathers with gray tips (Jensen et al., 2008). Phenotypic measurements were corrected for fieldworker variation by adding the mean difference between Thor Harald Ringsby measurement and a fieldworker measurement when this was significant (P<0.05) as judged by a paired t-test (see Kvalnes, 2016). When using one value per individual (mean phenotypic values), any variation in trait size because of age and season was accounted for by adjusting trait size to February-measures at the age of 1 year. This was done by first fitting a general linear mixed-effects model (using the lme4 package in R, Bates et al., 2015) for each trait and sex separately, with age, age2 and month as explanatory variables, and an individual random intercept and slope to separate out any between-individual variation in the relationship with age. The predicted values from this model were used to adjust each measurement of a trait through the life of an individual to its predicted value in February at the age of 1 year. Then, the mean of all adjusted measurements was used as an individual’s mean trait value (Kvalnes, 2016). We used this adjusted measurement as the mean trait estimate in all of the following analyses. The effects of sex, hatch year and hatch island were accounted for in the models below (heritability estimation, chromosome partitioning and GWAS) when these factors were significantly associated with the trait being analyzed (adjusted R2 and P-values in Supplementary Table S1). For the repeated measurements, we did not adjust trait measurements for age and season prior to the analyses, but accounted for the effects of sex, hatch year, hatch island, month and age of the individual at the time of measurement directly in the GWAS (adjusted R2 and P-values in Supplementary Table S2).
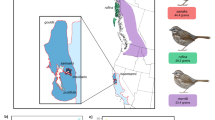
Schematics of phenotypic measurements in house sparrows (BL, bill length; BD, bill depth; TB, total badge; VB, visible badge; WL, wing length; TL, tarsus length) and in collared flycatchers (TL, tarsus length; WL, wing length; WWP, white wing patches). Photos by H Jensen (male house sparrow) and A Husby (male collared flycatcher).
Phenotypic data on collared flycatchers were collected from a nestbox population on the Swedish island of Öland (57°10’N, 16°58E), which has been monitored since 2002 (Qvarnström et al., 2010). Individuals were caught and ringed while breeding, or ringed as nestlings. For all adults, tarsus length, body mass, wing length and the size of white wing patches were measured. The white on the wing was measured using sliding calipers as the sum of the amount of white on primary feathers (two to seven). The effects of sex and study area were included in the models below (heritability estimation, chromosome partitioning and GWAS). Sex was included as a fixed effect in the mean models of body mass, wing length and white patches on the wings, whereas study area was included in the model of body mass and white patches on the wings (adjusted R2 and P-values in Supplementary Tables S3). For repeated measures models, sex was included as a covariate for body mass, wing length and white patches on the wings, and study area was included in models of tarsus, wing length and white on the wings. A description of the phenotypic data and number of records available for the analyses of house sparrows and collared flycatchers is reported in Table 2.
Genotyping
For both species, a small blood sample was taken from the brachial vein of each individual and stored in ethanol, Queens lysis buffer or FTA cards for subsequent DNA extraction. In total, we genotyped 1898 house sparrows with a 10K SNP array (Hagen et al., 2013) and 825 adult collared flycatchers on a 50K SNP array (Kawakami et al., 2014). We excluded markers with a call rate less than 95%, minor allele frequency of less than 0.01 and a P-value for rejection of Hardy–Weinberg equilibrium (HWE) of less than 0.001. We also excluded one of a pair of individuals in whom the identity by state was greater than 0.9 (removing accidental duplicated samples—for example, due to pipetting the same sample twice and avoiding bias errors introduced by these over-represented genotypes). For this quality-control step, we used the function check.marker() in GenABEL (Aulchenko et al., 2007). For house sparrows, the quality control for HWE was conducted independently for each population, and markers that failed this test in all populations were excluded (that is, when a marker was not at HWE in all populations, it was excluded). After quality control, 6348 SNPs were available for analysis in 1851 house sparrows and 38 689 SNPs for 825 collared flycatchers.
Genetic variance and heritability estimation
Three different software packages were used to estimate genetic variance and heritability (Table 1). We first estimated genomic heritability of the phenotypic traits using the R package RepeatABEL, using the function ‘rGLS’ (Rönnegård et al., 2016), which allows the use of repeated measurements of phenotypic traits when estimating genetic variance. We refer to the genomic heritability estimates from this approach as h2kin (rep). For comparison with other studies (Robinson et al., 2013; Santure et al., 2013), we also estimated heritability using the mean phenotype for each individual in the R package GenABEL, using the function ‘polygenic’ (Aulchenko et al., 2007). We refer to this estimate as h2kin (mean). Finally, we used the software GCTA to estimate the genetic variance using mean phenotypic values. In addition, GCTA was used to estimate the proportion of variance tagged by the SNP arrays (see below). In each of these methods, when appropriate (Supplementary Tables S1 and S2), we included various fixed effects. Ideally, some of the fixed effects would be included as random effects (hatch year and hatch island), but this was not possible because not all software allow more than one or two random effects (which are typically the relatedness matrices).
In addition to estimating genome-wide genetic variance, we also used a recent method to estimate how much of the genetic variance was captured by the SNP arrays (h2g). Unlike the method by Yang et al., 2010, which needs unrelated individuals, Zaitlen et al. (2013) use two genetic relationship matrices (GRMs) in a restricted maximum likelihood (REML) analysis to calculate both SNP-based heritability (h2g) and a pedigree-equivalent heritability (h2gkin) using all individuals. This method has been implemented in the software GCTA (Yang et al., 2011a).
The variance explained by all autosomal SNPs was estimated using the mixed-effects linear model  , where y is a vector of phenotypes, β is a vector of fixed effects (for example, sex, hatch island and hatch year) with its incidence matrix X, gG is a matrix of aggregate effects of all autosomal SNPs for all individuals and
, where y is a vector of phenotypes, β is a vector of fixed effects (for example, sex, hatch island and hatch year) with its incidence matrix X, gG is a matrix of aggregate effects of all autosomal SNPs for all individuals and  is a matrix of aggregate effects of all autosomal SNPs where unrelated individuals have off-diagonals that are <0.05 set to 0 to distinguish them from related individuals. This model therefore uses the mean phenotypic values and estimates additive genetic effects tagged by the genotyped SNPs (‘SNP-based heritability’—h2g) and the pedigree-equivalent heritability using information about genetic relationships of kin inferred from the marker data (‘total narrow-sense heritability’—h2gkin). The estimated total narrow-sense heritability (h2gkin) can therefore be compared to h2kin (mean) estimates from GenABEL (Zaitlen et al., 2013). Prediction errors due to imperfect linkage disequilibrium were adjusted using the —grm-adj 0 function when estimating genetic relationships (for similar approach, see Bérénos et al., 2015).
is a matrix of aggregate effects of all autosomal SNPs where unrelated individuals have off-diagonals that are <0.05 set to 0 to distinguish them from related individuals. This model therefore uses the mean phenotypic values and estimates additive genetic effects tagged by the genotyped SNPs (‘SNP-based heritability’—h2g) and the pedigree-equivalent heritability using information about genetic relationships of kin inferred from the marker data (‘total narrow-sense heritability’—h2gkin). The estimated total narrow-sense heritability (h2gkin) can therefore be compared to h2kin (mean) estimates from GenABEL (Zaitlen et al., 2013). Prediction errors due to imperfect linkage disequilibrium were adjusted using the —grm-adj 0 function when estimating genetic relationships (for similar approach, see Bérénos et al., 2015).
As sample size and marker density differ between species (1898 house sparrows genotyped on 6348 SNPs versus 825 collared flycatchers genotyped on 38 689 SNPs), this makes it difficult to compare heritability estimates. We therefore randomly downsampled the number of SNPs (in the collared flycatcher) and the number of individuals (in the house sparrows) across the data set such that both sample size and marker density were the same in both species. Heritabilities were then estimated using the four approaches described above and in Table 1.
Partitioning of genetic variance between chromosomes
To partition genetic variance among chromosomes, we used the GCTA software (Yang et al., 2011a) to compute chromosome-specific GRMs for the autosomes. The genetic variance attributable to each chromosome was estimated by fitting the GRMs of all chromosomes simultaneously in the model:  , where gc is a vector of genetic effects attributable to each chromosome with
, where gc is a vector of genetic effects attributable to each chromosome with  is the GRM from the SNPs on each chromosome and
is the GRM from the SNPs on each chromosome and  is the chromosome variance). The maximum number of chromosomes fitted differed between house sparrows (m=29) and collared flycatchers (m=33). If the models did not converge (for all traits except body mass in house sparrows and for wing length in collared flycatchers), the chromosomes with the smallest number of SNP markers were iteratively excluded until the model converged. A maximum of eight chromosomes were excluded for house sparrows and 10 chromosomes for collared flycatchers (Supplementary Tables S5 and S7).
is the chromosome variance). The maximum number of chromosomes fitted differed between house sparrows (m=29) and collared flycatchers (m=33). If the models did not converge (for all traits except body mass in house sparrows and for wing length in collared flycatchers), the chromosomes with the smallest number of SNP markers were iteratively excluded until the model converged. A maximum of eight chromosomes were excluded for house sparrows and 10 chromosomes for collared flycatchers (Supplementary Tables S5 and S7).
To address convergence problems, we first fitted separate models for each chromosome with the GRM of the focal chromosome and a GRM for all other chromosomes combined:  , where
, where  and Afocal is the GRM of the focal chromosome.
and Afocal is the GRM of the focal chromosome.  estimates the variation explained by all other chromosomes but the focal chromosome. However, this did not solve the convergence issues. We therefore also tried to estimate a single GRM using marker data from all microchromosomes jointly. This should estimate the variance due to all microchromosomes together. Unfortunately, this also did not completely solve the problem, and we still had some traits where the models did not converge (Supplementary Table S6). These convergence problems are likely because of the low number of markers on some chromosomes (the microchromosomes).
estimates the variation explained by all other chromosomes but the focal chromosome. However, this did not solve the convergence issues. We therefore also tried to estimate a single GRM using marker data from all microchromosomes jointly. This should estimate the variance due to all microchromosomes together. Unfortunately, this also did not completely solve the problem, and we still had some traits where the models did not converge (Supplementary Table S6). These convergence problems are likely because of the low number of markers on some chromosomes (the microchromosomes).
We estimated the proportion of variance explained by the Z chromosome in collared flycatchers compared to the proportion explained by all autosomes considered together (Supplementary Table S8). Note that we do not have comparable information in house sparrows (markers on the Z chromosome were not included here because they have not been mapped to the genome and a linkage map for the Z chromosome is not available yet), and so we did not consider this further.
To estimate the relationship between chromosome size and the amount of variation it explained for each trait, we used linear regression models in R (R Core Team, 2015). Chromosome sizes for both house sparrows and collared flycatchers were taken from the reference genome assemblies (house sparrows; NCBI accession number 17653, collared flycatchers; NCBI accession number 11872).
GWAS
Data on both house sparrows and collared flycatchers have been collected as part of long-term individual-based monitoring projects. This allowed us to take repeated measures of individuals and to take this nested data structure into account when testing for associations between SNPs and the phenotypic traits using the function ‘rGLS’ in the R package RepeatABEL (Rönnegård et al., 2016). RepeatABEL allows a mixed model with both a repeated measures effect as well as relatedness between individuals to be included as random effects. For comparison, we also used individual mean measurements of phenotypic traits using the function ‘grammar’ in the R package GenABEL (Aulchenko et al., 2007). Reported P-values are based on Wald tests and are corrected for population stratification (structure and relatedness) and the repeated sampling of the same individuals when using the repeated measures GWAS (Rönnegård et al., 2016). The genome-wide significance threshold was determined using a Bonferroni correction by dividing the significance value (P=0.05) by the number of markers (Lander and Kruglyak, 1995) resulting in P=7.80 × 10−6 for house sparrows and P=1.29 × 10−6 for collared flycatchers. This is a conservative P-value as it assumes that all markers are independent. We also report the additive genetic variance explained by each of the five SNPs with the smallest P-values, estimated as VSNP=2pqa2, where p and q are the frequencies of the major and minor allele frequencies, respectively, and a is the additive SNP effect (Falconer and Mackay, 1996). The heritability of each of these SNPs (h2SNP) was then estimated as VSNP/VP (where VP is the phenotypic variance estimate obtained from the GWAS).
Results
Genomic heritability
Heritability estimated using repeated phenotypic measures (h2kin (rep)) ranged from 0.136 for total badge size to 0.415 for tarsus length in house sparrows, and from 0.149 for white wing patches to 0.289 for tarsus length in collared flycatchers (Table 2). In general, when using the mean phenotypic values heritability estimates (h2kin (mean)) tended to be higher for both species across all traits, ranging from 0.228 for total badge size to 0.495 for bill length in house sparrows, and from 0.267 for white wing patches to 0.576 for tarsus length in collared flycatchers (Table 2).
When jointly estimating the SNP-based heritability (h2g) and total narrow-sense heritability (h2gkin) using the method by Zaitlen et al. (2013), we found that SNP-based heritability ranged from zero for wing length to 0.185 for body mass in house sparrows, and from 0.080 for body mass to 0.538 for wing length in collared flycatchers. For most traits, the total narrow-sense heritability estimates from the method by Zaitlen et al. (2013; Table 3) were similar to the h2kin(mean) values from GenABEL (Table 2).
In general, SNP-based heritabilities were higher for collared flycatchers compared with the house sparrows. To examine this in more detail, we thinned both data sets down to 825 individuals and 6348 SNPs. In house sparrows, the reduction of sample size caused an inflation for many h2 estimates and an increase in standard errors, whereas a reduction in marker density in collared flycatchers had little effect (Tables 4 and 5).
Chromosome partitioning
We found a significant linear relationship between the proportion of variance explained by each chromosome and chromosome size for tarsus length, body mass, bill length and visible badge size in house sparrows (Figure 2), but not for wing length, bill depth and total badge size (Figure 2 and Supplementary Table S5). The variance explained by each chromosome ranged from 0 to 0.092 across all traits (Supplementary Table S5). Chromosome 2, which is the largest chromosome in house sparrows, did not explain much of the variation for most traits, except for visible badge size. On the other hand, chromosome 1 (the second largest chromosome) explained a high proportion of the variance in most morphological traits (tarsus length, wing length, bill depth, bill length and total badge) except for body mass and visible badge size. Interestingly, a relatively small chromosome (14) explained a large proportion of the variation for wing length (Figure 2 and Supplementary Table S5).
Some models failed to convergence when including all chromosomes and estimating the variance using all microchromosomes together did not solve convergence issues (for example, wing length). These values were similar to estimates when fitting the GRMs of all chromosomes simultaneously in the model (Supplementary Tables S5 and S6). Fitting separate models with the GRM for a focal chromosome against the rest also did not solve convergence problems (see Materials and methods). Some of the chromosome-specific estimates should therefore be treated with caution.
For collared flycatchers, the relationship between the proportion of variance explained by each chromosome and chromosome size was significant for tarsus length (r=0.656), but not for wing length (r=0.269), body mass (r=0.144) or white wing patch (r=−0.041; Figure 3 and Supplementary Table S7). The proportion of variance explained by a single chromosome ranged from 0 to 0.150 across all traits. As with house sparrows, chromosome 2 did not explain substantial variation in any trait, whereas chromosome 1 contributed substantially to both tarsus and wing length. Chromosome 4 also contributed substantially to tarsus length (Supplementary Table S7), which reflects the presence of a significant marker for tarsus length on chromosome 4 (see below).
GWAS
After correcting for multiple testing, one SNP (11485) on chromosome 20 was significantly associated with bill length in house sparrows when using the mean phenotypic values, and explained 2% of the phenotypic variation (Supplementary Table S11 and Supplementary Figure S3). This SNP also had the lowest P-value when using repeated measures, although it was no longer significant after Bonferroni correction (Supplementary Table S9 and Supplementary Figure S1). In general, each one of the top five SNPs (ranked by P-value) explained only a small proportion of the phenotypic variation, and these values were similar between the two approaches. The total amount of variation explained by the top five SNPs ranged from 3% for wing length to 5.8% for total badge using the repeated measures, and from 3.3% for wing length to 10.9% for total badge using the mean values (Supplementary Tables S9 and S11). The ranking of the top five SNP associations was often similar between the two approaches, although they were not always shared (Supplementary Tables S9 and S11).
As for house sparrows, the results from the two GWAS in collared flycatchers were also concordant. In neither approach did we find significant associations between SNP markers and any phenotypic traits with the exception of tarsus length (SNP N00199:174262 on chromosome 4); this SNP explained a small amount of the variation (3% using repeated measures and 4% using mean values). Across all the traits measured, allelic variation at the top five SNPs was responsible for between 3.3 and 11.4% of the phenotypic variation (Supplementary Tables S9 and S12, Supplementary Figures S2 and S4).
Discussion
Understanding the genetic architecture of traits in wild populations can better elucidate the mechanisms responsible for trait evolution, including the expected rate of evolutionary change (Barton and Keightley, 2002). In this study, we used large-scale genotype data from custom SNP arrays from two passerine species to examine the genetic architecture of morphological traits. Using genomic data, we demonstrate that these traits are heritable (Tables 2 and 3), and chromosome partitioning revealed that for many traits the proportion of variance explained by a chromosome scaled with its size, suggesting a polygenic basis (Figures 2 and 3). This interpretation was further supported by the GWAS that did not detect any large-effect loci (Supplementary Tables S9–S12). Overall, our results add further support for a polygenic basis in morphological, sexually selected and life-history traits as earlier documented, for example, in great tit (Santure et al., 2013, 2015), Soay sheep (Bérénos et al., 2015) and collared flycatcher (Husby et al., 2015; Kardos et al., 2016).
The different approaches used here to estimate heritability gave similar values to that seen in previous studies using pedigree approaches in both species (Gustafsson, 1986; Jensen et al., 2003, 2008). As documented in a previous pedigree study (for example, Åkesson et al., 2008), the use of repeated measures resulted in somewhat lower estimates of heritability, and this was also the case for genomic heritability in the GWAS context (Rönnegård et al., 2016). Our results support this finding (Table 2), which is a result of reduced residual variance in the mean trait models. Our estimates of heritability were generally lower than the average heritability estimates for morphological traits in wild systems (Postma, 2014), which seems consistent with previous reports that genomic heritabilities tend to be lower than heritabilities estimated from pedigree-based animal models (Zaitlen et al., 2013; de los Campos et al., 2015).
A relatively new measure is the SNP-based heritability, which estimates how much of the variation in a trait is tagged by the SNP array used after accounting for the variance explained by similarity between relatives. Studies in humans have demonstrated that the SNP-based heritability is generally lower than the pedigree heritability (Yang et al., 2010), suggesting that not all causal sites are tagged by the SNP arrays used. We used a recent approach developed by Zaitlen et al. (2013) to simultaneously estimate the SNP-based heritability (h2g) and the total narrow-sense heritability (h2gkin). In general, and as expected, the SNP-based heritability tended to be lower than the total narrow-sense heritability (Tables 3 and 5). This could be because of a relatively low density of SNPs, compared with relatively many related individuals. Thus, we might estimate a higher heritability by using genomic relatedness to assess resemblance between relatives than by assessing the phenotypic variance explained by tagged SNPs. The SNP-based heritability in house sparrows was generally lower than that in collared flycatchers (0–0.185 versus 0.080–0.538, respectively), which was not unexpected, given that marker density is higher for collared flycatchers than house sparrows. These differences in SNP-based heritability may also be the result of house sparrows being more related than collared flycatchers as a consequence of their life-history characteristics.
We therefore thinned the collared flycatcher data set to have 6348 SNPs and the house sparrow data set to have 835 individuals. Interestingly, both h2gkin and h2(mean) showed that there was an upward bias in the heritability estimates in house sparrows compared with the full data set (Tables 4 and 5). This indicates that there may be an inflation of heritability estimates and effect sizes at lower biological sample sizes. We did not see a similar effect in flycatchers, where we thinned the number of markers. This could be because the number of markers in the full data set was already relatively low, particularly for the linkage disequilibrium structure typical in passerines (Kawakami et al., 2014; Kardos et al., 2016).
The method by Zaitlen et al. (2013) has not yet, to our knowledge, been used in other studies of natural populations. Surprisingly, the proportion of the heritability explained by the SNPs in our study is similar or higher to that seen in humans (Yang et al., 2010). However, these data sets have substantial differences in terms of SNP density, sample sizes, level of relatedness between individuals and chromosome architecture. As we have demonstrated by thinning the sparrow data set to fewer individuals, there may be an inflation of the amount of phenotypic variation explained by kinship-based methods when fewer individuals are included in an analysis. Simulations are needed for a robust comparison and to understand the effects of these differences in the data set when estimating SNP-based heritabilities.
For some traits, chromosome-partitioning analyses demonstrated a significant positive association between the amount of variation explained by a chromosome and the size of that chromosome, as would be expected if the trait was polygenic (Figures 2 and 3). However, we did not find significant correlation between chromosome size and proportion of variance explained for wing length, bill depth and total badge in house sparrows, or wing length, body mass and white wing patches in collared flycatchers, although some larger chromosomes explained substantial amounts of the overall variation (Figures 2 and 3). Similar morphological traits have been identified as polygenic in other species—for example, wing length, weight, tarsus length, clutch size and egg weight in great tits (Robinson et al., 2013; Santure et al., 2015) and jaw size and body mass in Soay sheep (Bérénos et al., 2015). In addition, Schielzeth et al. (2012) used a QTL linkage mapping approach to find six genomic regions linked to variation in wing length in a captive population of zebra finches. All putative regions showed similar effect sizes (3.9–8.3%) and together explained only about half of the heritability in wing length. The many candidate genes within the QTL regions further suggest a polygenic basis for wing length in zebra finches. In total, it seems that these morphological traits in passerines could generally be polygenic.
One may still argue that our results are not totally consistent with previous findings. However, it is important to keep in mind that the larger chromosomes tended to explain substantial variance also in traits that did not show significant correlation between chromosome size and proportion of variance explained (Supplementary Tables S5–S7), as expected under a polygenic model. Moreover, estimating relatedness on the microchromosomes is difficult because we have very few markers on these, which makes estimation difficult (and potentially unreliable), as indicated by the problems with model convergence. An additional consideration is that it is not clear that chromosomes that contribute disproportionately to trait variation, given their size should harbor large-effect QTLs because it is equally plausible that many small-effect loci cluster on that chromosome. As pointed out by Schielzeth and Husby (2014), such clustering of many loci of small effect on a single chromosome is not uncommon and can involve association with biologically relevant pathways for a specific trait. Some caution is, therefore, warranted when making predictions about the genetic architecture of traits from regressions of chromosome size on proportion variance explained. Finally, we did not find any significant single large-effect-size markers for these traits on the chromosomes that explained a disproportional part of the variance. Taken together, most evidence, therefore, points in the direction of a polygenic basis also for these traits.
We only detected two SNP markers that met the genome-wide significant threshold: one SNP on chromosome 20 for bill morphology in house sparrows that explained 1.9% of the phenotypic variation and one SNP on chromosome 4 in the collared flycatcher for tarsus length explaining 3% of the variation. SNP 11485 on chromosome 20 associated with bill length in house sparrows might be related to a previously detected QTL on chromosome 20 for beak morphology in zebra finches (Knief et al., 2012). In the zebra finch, this QTL was found to be located at 0.86–14.17 Mb, and the position of the SNP in our study is 7.6 Mb. We are not aware of any previous studies on tarsus length that show an association in the region on chromosome 4 where the QTL for tarsus length in the collared flycatcher was located.
Another interesting finding in our study was the presence of two shared SNPs for different traits in house sparrows. SNP 15053 was among the top five SNPs associated with both total badge and visible badge size when using repeated values (Supplementary Table S9), and SNP 11485 was among the top five SNPs associated with bill depth and bill length when using the mean values (Supplementary Table S11). Shared loci among traits will result in a genetic correlation between these traits (that is, total badge versus visible badge size and bill depth versus bill length), and these are traits that have previously been found to be genetically correlated in this species (Jensen et al., 2008).
In summary, we genotyped a large number of individual house sparrows and collared flycatchers on custom genome-wide SNP arrays and examined the genetic architecture of a number of phenotypic traits. By estimating and using kinship matrices based on genome-wide SNP data, we demonstrated that all traits showed substantial amount of genetic variance, in line with results from previous pedigree-based approaches. When applying a novel method to estimate the proportion of variance in the traits captured by the genotyped SNPs (SNP-based heritability, h2g), our estimates were somewhat larger than those expected considering the sample size and number of SNPs used. The SNP-based heritability was lower than the total narrow-sense heritability in both species, suggesting that not all causal sites are tagged by the SNP arrays used. Chromosome partitioning as well as GWAS showed several lines of evidence, suggesting that the investigated traits are polygenic. This was indicated by a positive correlation between chromosome size and amount of variance explained for most traits, a lack of any large-effect QTLs and the small amount of total variation explained by the top SNPs in the GWAS. Our results are in line with other recent studies showing a polygenic basis to phenotypic traits in natural populations.
Finally, one major conclusion to make from this work is that genomic techniques, even with low marker densities, can be useful to provide a better understanding of short-term evolutionary change of phenotypic traits in natural populations. We are currently transitioning to studies at the level of entire genomes but low-density SNP arrays can be very useful tools. In particular, these SNP arrays are a cost-efficient resource for addressing questions that require large sample sizes from natural populations.
Data Archiving
Data are available from the Dryad Digital Repository http://dx.doi.org/10.5061/dryad.786m4. The deposit files contain data for each sample with the locality and genotypes for all individuals.
References
Åkesson M, Bensch S, Hasselquist D, Tarka M, Hansson B . (2008). Estimating heritabilities and genetic correlations: Comparing the “animal model” with parent-offspring regression using data from a natural population. PLoS ONE 3: e1739.
Anderson TR . (2006) Biology of the Ubiquitous House Sparrow: from Genes to Populations. Oxford University Press: New York, NY, USA.
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM . (2007). GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296.
Barson NJ, Aykanat T, Hindar K, Baranski M, Bolstad GH, Fiske P et al. (2015). Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 528: 405–408.
Barton NH, Keightley PD . (2002). Understanding quantitative genetic variation. Nat Rev Genet 3: 11–21.
Bates D, Mächler M, Bolker BM, Walker SC . (2015). Fitting linear mixed-effects models using lme4. J Stat Software 67: 1–48.
Bérénos C, Ellis PA, Pilkington JG, Lee SH, Gratten J, Pemberton JM . (2015). Heterogeneity of genetic architecture of body size traits in a free-living population. Mol Ecol 24: 1810–1830.
Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T et al. (2012). The genomic landscape of species divergence in Ficedula flycatchers. Nature 491: 756–760.
Falconer D, Mackay T . (1996) Introduction to Quantitative Genetics. 4th edn. Longmans Green, Harlow: Essex, UK.
Goddard ME, Hayes BJ . (2009). Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet 10: 381–391.
Gustafsson L . (1986). Lifetime reproductive success and heritability: empirical support for fisher’s fundamental theorem. Am Nat 128: 761–764.
Hagen IJ, Billing AM, Rønning B, Pedersen SA, Pärn H, Slate J et al. (2013). The easy road to genome-wide medium density SNP screening in a non-model species: development and application of a 10 K SNP-chip for the house sparrow (Passer domesticus. Mol Ecol Resources 13: 429–439.
Husby A, Kawakami T, Ronnegard L, Ellegren H, Qvarnstrom A . (2015). Genome-wide association mapping in a wild avian population identifies a link between genetic and phenotypic variation in a life-history trait. Proc R Soc B 282: 20150156.
Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C et al. (2014). Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346: 1126–1138.
Jensen H, Sæther BE, Ringsby TH, Tufto J, Griffith SC, Ellegren H . (2003). Sexual variation in heritability and genetic correlations of morphological traits in house sparrow (Passer domesticus. J Evol Biol 16: 1296–1307.
Jensen H, Steinsland I, Ringsby TH, Sæther BE . (2008). Evolutionary dynamics of a sexual ornament in the house sparrow (Passer domesticus: the role of indirect selection within and between sexes. Evolution 62: 1275–1293.
Johnston SE, Orell P, Pritchard VL, Kent MP, Lien S, Niemelä E et al. (2014). Genome-wide SNP analysis reveals a genetic basis for sea-age variation in a wild population of Atlantic salmon (Salmo salar. Mol Ecol 23: 3452–3468.
Kardos M, Husby A, Mcfarlane SE, Qvarnström A, Ellegren H . (2016). Whole-genome resequencing of extreme phenotypes in collared flycatchers highlights the difficulty of detecting quantitative trait loci in natural populations. Mol Ecol Resources 16: 727–741.
Kawakami T, Backstrom N, Burri R, Husby A, Olason P, Rice AM et al. (2014). Estimation of linkage disequilibrium and interspecific gene flow in Ficedula flycatchers by a newly developed 50k single-nucleotide polymorphism array. Mol Ecol Resources 14: 1248–1260.
Knief U, Schielzeth H, Kempenaers B, Ellegren H, Forstmeier W . (2012). QTL and quantitative genetic analysis of beak morphology reveals patterns of standing genetic variation in an Estrildid finch. Mol Ecol 21: 3704–3717.
Kvalnes T . (2016) Evolution by natural selection in age-structured populations in fluctuating environments. PhD thesis, Norwegian University of Science and Technology, Norway.
Lamichhaney S, Han F, Berglund J, Wang C, Almen M, Webster M et al. (2016). A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 352: 470–474.
Lander E, Kruglyak L . (1995). Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11: 241–247.
de los Campos G, Sorensen D, Gianola D . (2015). Genomic heritability: what is it? PLoS Genet 11: e1005048.
Lynch M, Walsh B . (1998) Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland, MA, USA.
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al. (2009). Finding the missing heritability of complex diseases. Nature 461: 747–753.
Postma E (2014). Four decades of estimating heritabilities in wild vertebrate populations: improved methods, more data, better estimates? In: Charmantier A, Garan D, Kruuk L (eds). Quantitative Genetics in the Wild. Oxford University Press: Oxford, UK, pp 16–39..
Qvarnström A, Rice AM, Ellegren H . (2010). Speciation in Ficedula flycatchers. Philos Trans R Soc B BiolSci 365: 1841–1852.
R Core Team. (2015) R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Robinson MR, Santure AW, Decauwer I, Sheldon BC, Slate J . (2013). Partitioning of genetic variation across the genome using multimarker methods in a wild bird population. Mol Ecol 22: 3963–3980.
Rönnegård L, McFarlane SE, Husby A, Kawakami T, Ellegren H, Qvarnström A . (2016). Increasing the power of genome wide association studies in natural populations using repeated measures-evaluation and implementation. Methods Ecol Evol 7: 792–799.
Santure AW, De Cauwer I, Robinson MR, Poissant J, Sheldon BC, Slate J . (2013). Genomic dissection of variation in clutch size and egg mass in a wild great tit (Parus major population. Mol Ecol 22: 3949–3962.
Santure AW, Poissant J, De Cauwer I, van Oers K, Robinson MR, Quinn JL et al. (2015). Replicated analysis of the genetic architecture of quantitative traits in two wild great tit populations. Mol Ecol 24: 6148–6162.
Schielzeth H, Forstmeier W, Kempenaers B, Ellegren H . (2012). QTL linkage mapping of wing length in zebra finch using genome-wide single nucleotide polymorphisms markers. Mol Ecol 21: 329–339.
Schielzeth H, Husby A . (2014). Challenges and prospects in genome-wide quantitative trait loci mapping of standing genetic variation in natural populations. Ann N Y Acad Sci 1320: 35–57.
Slate J, Santure AW, Feulner PGD, Brown EA, Ball AD, Johnston SE et al. (2010). Genome mapping in intensively studied wild vertebrate populations. Trends Genet 26: 275–284.
Veiga J . (1996). Permanent exposure versus facultative concealment of sexual traits: an experimental study in the house sparrow. Behav Ecol Sociobiol 39: 345–352.
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR et al. (2010). Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569.
Yang J, Lee SH, Goddard ME, Visscher PM . (2011a). GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82.
Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM et al. (2011b). Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet 43: 519–525.
Zaitlen N, Kraft P, Patterson N, Pasaniuc B, Bhatia G, Pollack S et al. (2013). Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet 9: e1003520.
Acknowledgements
We thank students and fieldworkers for their contribution in the fieldwork, and students and laboratory technicians for help with the laboratory work. We are also grateful to the inhabitants in the study areas, whose hospitality made this study possible. The research done on house sparrows was carried out in accordance with permits from the Norwegian Animal Research Authority and the Ringing Centre at Stavanger Museum, Norway. The research on collared flycatchers was done with permission from the Linköping animal ethics committee. This study was supported by grants from the Research Council of Norway (191847, 204303, 221956 and 214553 to AH, HJ, BE-S) and its Centre of Excellence scheme (223257), the Swedish Research Council (AQ, HE), the Knut and Alice Wallenberg Foundation (HE), the European Research Council (HE) and Natural Sciences and Engineering Research Council of Canada (SEM). The 10K SNP genotyping of house sparrows was provided by the Genomics Core Facility, Norwegian University of Science and Technology. Genotyping of collared flycatchers was performed by the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University. We also thank the associate editor, Anna Santure, and two anonymous reviewers for comments that greatly improved the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Silva, C., McFarlane, S., Hagen, I. et al. Insights into the genetic architecture of morphological traits in two passerine bird species. Heredity 119, 197–205 (2017). https://doi.org/10.1038/hdy.2017.29
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2017.29
This article is cited by
-
Investigating pedigree- and SNP-associated components of heritability in a wild population of Soay sheep
Heredity (2024)
-
The effect of repeated measurements and within-individual variance on the estimation of heritability: a simulation study
Behavioral Ecology and Sociobiology (2024)
-
Decomposing phenotypic skew and its effects on the predicted response to strong selection
Nature Ecology & Evolution (2022)
-
Linking genetic, morphological, and behavioural divergence between inland island and mainland deer mice
Heredity (2022)
-
Animal models with group-specific additive genetic variances: extending genetic group models
Genetics Selection Evolution (2019)