Abstract
Chronic traumatic encephalopathy (CTE) is a progressive neurodegenerative disorder that is associated with repetitive head injury and has distinctive neuropathological features that differentiate this disease from other neurodegenerative diseases. Intraneuronal tau aggregates, although they occur in different patterns, are diagnostic neuropathological features of CTE, but the precise mechanism of tauopathy is not known in CTE. We performed whole RNA sequencing analysis of post-mortem brain tissue from patients with CTE and compared the results to normal controls to determine the transcriptome signature changes associated with CTE. The results showed that the genes related to the MAP kinase and calcium-signaling pathways were significantly downregulated in CTE. The altered expression of protein phosphatases (PPs) in these networks further suggested that the tauopathy observed in CTE involves common pathological mechanisms similar to Alzheimer’s disease (AD). Using cell lines and animal models, we also showed that reduced PPP3CA/PP2B phosphatase activity is directly associated with increases in phosphorylated (p)-tau proteins. These findings provide important insights into PP-dependent neurodegeneration and may lead to novel therapeutic approaches to reduce the tauopathy associated with CTE.
Similar content being viewed by others
Introduction
Concussion, which is the most common neurological form of traumatic brain injury (TBI), occurs frequently in sports. Approximately 1.6–3.8 million sports-related concussions are reported annually in the United States.1, 2, 3, 4 Chronic traumatic encephalopathy (CTE) is a progressive brain disorder that is associated with repetitive brain trauma, including concussions and asymptomatic subconcussive head injury.5, 6 The clinical symptoms of CTE are aggression, depression, impulsivity, irritability, memory disturbances and increased suicidality.4 Although the clinical symptoms of CTE were described in the 1920s,7 neuropathologically confirmed CTE has been reported more recently in retired professional football players and other contact sport athletes with a history of repetitive head trauma.4, 8, 9, 10, 11, 12 The neuropathology of CTE is defined by the distinct perivascular accumulation of hyperphosphorylated tau in neurons and astrocytes at the depths of cerebral sulci.12 There is also evidence that the tauopathy spreads with age and the increasing severity of CTE to involve widespread regions of the brain. This progression of pathological severity has been divided into four stages of advancing severity, stages I through IV.11 Other pathologies associated with CTE include axonal injury, neuroinflammation and the deposition of TDP43.11 Clinical symptoms of CTE include behavior and mood changes, memory loss and cognitive decline that may, in some cases, evolve to overt dementia.9, 11, 13, 14 There is typically a latent period of years to decades between exposure to brain injury and the development of clinical symptoms.
Although the diagnosis and monitoring of CTE have improved over the last decade, a potential contribution of altered gene expression to the pathogenesis of CTE has not yet been studied. In this context, to identify and characterize the transcriptome signatures associated with the pathophysiology of CTE, we performed whole genome-wide RNA sequencing analysis of post-mortem brain tissue from patients with CTE and normal subjects. We further applied an in vitro cell line model and in vivo mouse model to validate how transcriptome changes lead to neuropathological changes in CTE. Since CTE and AD show a similar pathology in terms of increased tau hyperphosphorylation and tauopathy, we analyzed whether altered transcriptome signatures are associated with the tau phosphorylation pathway in CTE and AD. In addition, we verified the immunoreactivity of p-Tau in the post-mortem brain tissue of CTE and AD.
Materials and methods
Human tissues
Neuropathological processing of control, AD and CTE human brain samples was performed according to the procedures previously established for the Boston University Alzheimer’s Disease Center (BUADC) and Chronic Traumatic Encephalopathy (CTE) Center. Institutional review board approval for ethical permission was obtained through the BUADC and CTE Center. This study was reviewed by the Boston University School of Medicine Institutional Review Board (Protocol H-28974) and was approved as exempt because the study involved only tissue collected from post-mortem individuals, which are not classified as human subjects. Next of kin provided informed consent for participation and brain donation. The study was performed in accordance with the institutional regulatory guidelines and principles of human subject protection in the Declaration of Helsinki. Detailed information about the brain tissues is described in Supplementary Tables 1 and 2. In all cases in which AD was diagnosed at autopsy, AD was stated as the cause of death.
RNA sequencing and analysis
The samples were prepared for sequencing using the Illumina TruSeq RNA sample preparation kit according to the manufacturer’s instructions and sequenced on a HiSeq 2000 platform (Illumina, San Diego, CA, USA). The 101-bp sequenced paired-end reads were mapped to the hg19 reference human genome using the STAR 2-pass method.15 We used HTSeq to count the reads aligned to each gene based on the Ensembl gene set16 (Supplementary Table 3). We excluded samples that failed in the library preparation or sequence process. We also excluded samples with fewer than 10 million reads sequenced. Overall, 18 CTE subjects and 24 normal subjects were examined. The normalized read counts were applied to principal component analysis or clustering analysis, which was conducted through R and Cluster 3.0 and visualized via Java Treeview.16, 17, 18
Weighted gene co-expression network analysis
Co-expression analysis was performed using the weighted gene co-expression network analysis (WGCNA) method via its R package (v3.0.0).19 Overall, 24 682 genes were used to construct each network. To construct the networks, the absolute values of Pearson’s correlation coefficients were calculated for all possible gene pairs, and the resulting values were transformed such that the final matrix followed an approximate scale-free topology. A connectivity measure (k) per gene was calculated by summing the connection strengths with other genes. Modules are defined as clusters of densely interconnected genes; by default, the modules are indicated by branches of a hierarchical clustering tree using a dissimilarity measure. Each module is subsequently assigned a color. The gene expression profiles of each module were summarized according to the module eigengene (defined as the first principal component of the module expression levels). A measure of gene significance (GS) was computed to evaluate a gene’s correlation with a phenotype. The intra-modular connectivity (k-within) was calculated for each gene by summing the connection strengths with other module genes and dividing this number by the maximum intra-modular connectivity. The adjacency threshold for including edges in the output was set at 0.2. The eigengene-based connectivity of a gene in a module is defined as the correlation of the gene with the corresponding module eigengene.
Enrichment analyses based on Kyoto Encyclopedia of Genes and Genomes
Functional annotation analysis was used to assign biological relevance to the gene network modules using Gene Set Enrichment Analysis (GSEA) with Kyoto Encyclopedia of Genes and Genome (KEGG) pathway.20 The enrichment of the KEGG terms in each module was evaluated based on the hypergeometric test. The output from GSEA is an enrichment score that describes the imbalance in the distribution of ranks of gene expression in each gene set. The number of genes in the overlap (k) was set at ≥3. The enrichment score was normalized according to the size of the gene sets, which were subsequently ranked according to the normalized enrichment score. The default false-positive rate (FDR) q-value setting (FDR q-value <0.05) was used as the cut-off.
Tau BiFC cell lines and culture
The establishment of the HEK293/tau-BiFC cell line was previously described.21 Venus fluorescence protein amino-terminal fragment (VN 173) and carboxyl-terminal fragment (VC 155), which are independently non-fluorescent, were fused to full-length tau proteins. For neuronal cell expression, tau-BiFC constructs (pCMV6-hTau40-VN173 and pCMV6-hTau-VC155) were transfected into SH-SY5Y. SH-SY5Y/tau-BiFC stable cells were selected using Geneticin (200 μg ml−1). The phosphorylation of tau resulted in the assembly, or aggregation, of two tau molecules, which enabled the maturation of the Venus protein to emit a fluorescence signal. All established cell lines were grown in Dulbecco’s modified eagle medium containing 10% fetal bovine serum, 10 000 units per ml penicillin, 10 000 μg ml−1 streptomycin and 1 μg ml−1 of geneticin at 37 °C in a humidified atmosphere containing 5% CO2.
Live cell imaging and analysis
For microscopic image analysis, HEK293 tau-BiFC and SH-SY5Y tau-BiFC cells were plated onto a black 384-well plate. The next day, tau-BiFC cells were treated with okadaic acid (O8010, Sigma, St Louis, MO, USA), cyclosporine A (C3662, Sigma) or deltamethrin (D9315, Sigma) at various concentrations. After incubation for 29 h, the plate was imaged using the Operetta high contents imaging system (PerkinElmer). The cellular intensities of tau-BiFC fluorescence were analyzed using the Harmony 3.1 analysis software. Each experiment was performed in triplicate. The bar graphs for BiFC intensity indicate the means±s.e.m. from three independent experiments.
Confocal microscopy
Tissues and cells were immunostained for p-Tau (S199) (rabbit polyclonal, 1:200; Abcam, Cambridge, UK), p-Tau (S202/T205) (mouse monoclonal, 1:500; Thermo Scientific, Waltham, MA, USA) and PP2B/PPP3CA (rabbit polyclonal, 1:200; SantaCruz Biotech, Dallas, TX, USA) according to a previous report.22 For confocal microscopy, the specimens were incubated for 1 h with fluorescence (FITC)-conjugated secondary antibody (Vector, Burlingame, CA, USA) and Cy3-conjugated secondary antibody (Jackson Lab, West Grove, PA, USA) after the primary antibody incubation. The images were analyzed using a Spinning Disk Confocal microscope (IX2-DSU, Olympus, Tokyo, Japan). Pre-absorption with excess target protein or omission of primary antibody was used to demonstrate antibody specificity and determine the background generated from the detection assay.
Immunohistochemistry
Immunohistochemistry was performed as previously described.23 Paraffin-embedded tissues were sectioned in a coronal plane at 10–20 μm. The tissue sections were rehydrated, blocked with blocking solution (1% H2O2), and incubated with p-Tau (S202/T205) (1:200), PP2B/PPP3CA (1:200; SantaCruz Biotech), p-Tau (S199) (1:200; Abcam) and anti-III tubulin antibody (1:500 dilutions; Sigma) for 24 h. After washing three times, the slides were processed with Vector ABC Kit (Vector Lab). The immunoreactive signals were developed with DAB chromogen (Thermo Fisher Scientific, Meridian, Rockford, IL, USA) and analyzed under a bright field microscope.
Western blot analysis
Western blot analysis was performed as previously described.24 For the detection of p-Tau and other proteins, the blots were probed with anti-p-Tau (S199) (1:1000; Abcam), anti-p-Tau (S396) (1:1000), anti-p-Tau (AT8, S202/T205) (1:1000) and anti-β-actin (1:10 000; Sigma Aldrich, St Louis, MO, USA) antibodies, followed by treatment with the appropriate secondary antibodies conjugated to horseradish peroxidase (Pierce, 170-6515 and 170-6516). Immunoreactivity was detected using an enhanced chemiluminescence (ECL) kit (Thermo Scientific).
Quantitative real-time PCR
Total RNA was extracted from the frozen brain tissues using TRIzol reagent (MRC, TR118) as previously described (Lee et al.).23 Fifty nanograms of RNA was used as a template for quantitative RT-PCR amplification, using SYBR Green Real-time PCR Master Mix (Toyobo, QPK-201, Osaka, Japan). The primers were standardized in the linear range of the cycle prior to the onset of the plateau. The primer sequences are shown in Supplementary Table 4. GAPDH was used as an internal control. Real-time data acquisition was performed using an LightCyler96 Real-Time PCR System (Roche Diagnostics, Indianapolis, IN, USA) under the following cycling conditions: 95 °C for 1 min × 1 cycle, and 95 °C for 15 s, followed by 60 °C for 1 min × 45 cycles. The relative gene expression was analyzed using the LightCyler96 software and expressed as Ct the number of cycles needed to generate a fluorescent signal above a predefined threshold.
Construction and delivery of rAAV vectors containing PPP3CA shRNA
For in vivo gene silencing, the validated mouse shRNA sequences for PPP3CA were cloned into the pSicoR vector using HpaI/XhoI sites (Addgene, Cambridge, MA, USA, #21907) and subcloned into the pAAV-MCS vector (Stratagene) using MluI/BglII sites. The high-titer rAAV vectors were produced in HEK293TN cells using a helper virus-free system. Briefly, the rAAV vectors were produced after co-transfecting with equimolar amounts of a rep/cap/helper plasmid. After incubation for 72 h, the cells were lysed, treated with benzonase (Sigma #E1014) and further purified using HiTrap heparin columns (GE Healthcare, Pittsburg, CA, USA, #17-0460-01). Amicon ultra-15 centrifugal filter units (Millipore, Billerica, MA, USA #UFC9100) were used to concentrate up to the final volume. For the delivery of rAAV-PPP3CA shRNA into the brain, the stereotactic microinjection method was used. rAAV-shControl and AAV-PPP3CA shRNA were delivered into the dentate gyrus (AP: −2, ML: 1.5, DV: −1.85) of 4-month-old wild-type mice and Tau transgenic (P301L) mice as previously described.
Animal model of TBI
All procedures for the animal study were approved by Institutional Animal Care and Use Committee (IACUC) in the Korea Institute of Science and Technology according to international standards and guidelines. Wild-type (C57BL/6) and Tau transgenic (P301L) mice at 4 months of age were used in this study. The mice were fed a standard laboratory diet and water ad libitum in a controlled environment. The mice received three weight-drop-induced closed diffuse TBIs at 3-day intervals. We induced closed diffuse TBI by using a weight-drop device (weight 100 g, fall height 75 cm, angle 90°) as described previously but with a slight modification.25 The anatomical locus of impact was adjusted to bregma −1 to ±4. All mice were initially anesthetized with 2% avertin (23 μg g−1) IP injection before receiving the weight-drop-induced TBI. Sham-injured animals were subjected to the same protocol of anesthesia administration, but no mass was ever dropped. After TBI, the mice were placed in the supine position in a clean cage heated using a commercially available heating pad. The mice were then returned to their home cages after normal behavior (for example, grooming, walking, exploring) was recovered. We examined animal behaviors based on open field and assessed neurohistological changes. The animals were killed at 24 h after the last impact, and sham-operated animals were killed at 24 h after the last anesthesia.
Results
Transcriptome analysis of CTE
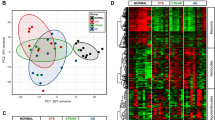
The potential contribution of altered gene expression to the pathogenesis of CTE has not been previously studied. We performed RNA sequencing on extracted RNA from four regions of post-mortem brain (anterior temporal (AT), posterior visual (PV), superior frontal (SF) and superior parietal (SP) cortex) from six subjects with neuropathologically verified CTE stage III and stage IV and seven normal controls (Supplementary Table 1). Paired-end RNA-Seq was performed using the Illumina HiSeq 2000 platform, and sequencing reads were aligned to the hg19 reference human genome. The average number of reads was ~71 million (74 million for normal subjects and 76 million for CTE subjects), with ~77.4% of the total reads mapping to the human transcriptome (Supplementary Table 5). To assess the similarities and differences among samples, we conducted a principal component analysis for the RNA-seq samples. The results demonstrated a clear distinction in the transcriptomic landscape between CTE and normal subjects (Figure 1a).
Global gene networks. (a) Principal component analysis (PCA) of gene expression profiles of all samples. CTE subjects are shown in blue and normal subjects are shown in red. (b) Dendrogram from gene coexpression network analysis of human CTE and normal subjects. Modules of co-expressed genes were assigned colors, as indicated by the horizontal bars under the dendrogram. Each line is a single gene where height (y axis) indicates dissimilarity. In total, 12 modules ranging from 74 to 3355 genes in size were identified. The 13 331 genes that were not co-expressed in the data set were assigned to the gray group. (c) Matrix with the Module-Trait Relationships (MTRs) and corresponding P-values between the detected modules on the x axis and selected CTE and brain-specific regions (AT, PV, SF and SP) on the y axis. The Pearson’s correlation coefficient (PCC) values range from −1 to +1 depending on the strength of the relationship. A positive value indicates that the transcripts within a particular co-expression module increase as the variable increases, whereas the opposite is true if the sign of the PCC is negative. Each PCC value is accompanied by the corresponding P-value in brackets. (d) Gene set enrichment analysis with Kyoto Encyclopedia of Genes and Genomes pathways using genes in seven modules with a high correlation to CTE. A maximum of 2000 high-module membership genes were used.
Gene network analysis identifies co-expressed gene modules
We applied a WGCNA19 to identify CTE-associated co-expression modules and their key constituents. WGCNA clusters genes with similar expression patterns in an unbiased manner, thus enabling the biological interpretation of transcriptional patterns. We identified 12 distinct co-expression modules, which varied in size from 74 to 3355 genes. A total of 13 331 genes did not share similar co-expression with the other genes in the network and were thus not included in any of the modules (Figure 1b,Supplementary Table 6). The module eigengene represented the expression of all genes classified into that module. When each eigengene expression was correlated against CTE status or other experimental variables, such as brain region (AT, PV, SF and SP) using Pearson’s method, only CTE status significantly correlated positively or negatively with a module eigengenes (Figure 1c). The blue module (r=−0.87, P=8 × 10−14) was negatively correlated with CTE subjects, indicating that most of genes in this module were downregulated in the CTE brain, whereas other significant modules for CTE were positively correlated. The results regarding the changing expression patterns of cell type-specific genes suggest that the genes related to both neurons and oligodendrocytes are affected by CTE progression. Specifically, neuron-associated genes were enriched in the blue module and showed decreased expression with CTE (Supplementary Figure 1). Consistent with the principal component analysis, the WGCNA analysis revealed no significant differences in the four brain regions. Only AT and PV were positively correlated with purple and black modules, respectively.
To understand the biological relevance of the identified modules highly correlated to CTE, we performed gene set enrichment analysis (GSEA) with KEGG pathways (Figure 1d). Two (blue and turquoise modules) of the seven modules were highly enriched for specific functional pathways that are directly associated with neurodegenerative diseases and neurobiological functions. In particular, the blue module was enriched for functional categories explicitly related to the calcium-signaling pathway, Alzheimer’s disease, Huntington’s disease and Parkinson’s disease and to genes belonging to the MAPK-signaling pathway. The brown and black modules were enriched for genes belonging to pathways involved in cancer, focal adhesion and actin cytoskeleton regulation. This analysis suggests that the large coordinated changes in gene expression are specifically associated with CTE brain.
The blue gene co-expression module is significantly associated with CTE
The blue module contains 3064 genes with a high correlation between the members of its module and gene significance for CTE (r=0.85 and P<1 × 10−200, Supplementary Figure 2a). Unsupervised hierarchical clustering performed on gene expression levels of the blue module showed the distinct separation of CTE from normal subjects (Supplementary Figure 2b). Eight hundred and seventy-two (872) upregulated and 2192 downregulated genes were abnormally expressed in CTE. Functional annotation analyses based on KEGG pathways using upregulated and downregulated genes in the blue module are provided in Figure 2a. The upregulated genes were primarily involved in the MAPK pathway, which is associated with Alzheimer’s disease pathogenesis. However, the tau phosphorylation kinase, p38 MAPK (MAPK11, MAPK12, MAPK13 and MAPK14), was not activated or associated with the blue module. Moreover, the expression levels of the other tau phosphorylation kinases, GSK3 (GSK3A and GSK3B), CDK5 and FYN, were not also increased in CTE (Supplementary Figure 2c). The downregulated genes were associated with several neurodegenerative diseases. Among these genes, we focused on genes not associated with Huntington’s disease and Parkinson’s disease, which do not have tauopathy for main disease progression. Using this approach, we identified nine genes that might affect CTE progression: CACNA1F, ATP2A2, GRIN2A, PPP3CA, PPP3CB, PPP3R1, CALM1, CALM2, CALM3 and CALML3. From the above-mentioned genes, we identified PPP3CA, encoding a calcium-dependent, calmodulin-stimulated protein phosphatase (PP), which was reported to be decreased in the medial temporal gyrus from AD patient brains.26 In addition, PPP3CB and PPP3R1, which are subunits of PP2B, were significantly decreased in CTE subjects (Figure 2b).
Reduction of PPP3CA/PP2B activity is associated with tauopathy in CTE
We explored whether altered PPP3CA gene expression also contributes to pathological tau phosphorylation and tauopathy in CTE. First, the downregulation of PPP3CA was confirmed through quantitative real-time PCR (qPCR) in the cortex of human CTE and normal subjects (Figure 3a). Western blot and densitometry analyses subsequently showed that the levels of PPP3CA were markedly decreased in CTE compared to normal subjects (Figure 3b and c). Furthermore, the levels of p-tau at S199, S202/T205 and S396, which are the most common sites of tau phosphorylation in neurodegenerative disorders, were increased 3- and 30-fold in CTE. Importantly, the levels of PPP3CA protein and p-tau (S202/T205) in the frontal cortex of CTE patients were inversely correlated. To further investigate the pathological characteristics of tauopathy in CTE, we performed 3,3′-diaminobenzidine (DAB) immunohistochemical staining in the cortex of normal and CTE brains (Figure 3d and e). Consistent with the qPCR and western blot data, the immunoreactivity of PPP3CA was decreased, and tau was hyperphosphorylated (S199, S396, S205/T208) in CTE.
Deregulation of PPP3CA is associated with the hyperphosphorylation of tau in chronic traumatic encephalopathy (CTE). (a) The mRNA levels of PPP3CA and PPP3CB are reduced in CTE (n=11) compared to normal subjects (n=11). (b) Protein levels of PPP3CA and PPP3CB are reduced but levels of p-tau (S199, S202/T205 and S396) are elevated in CTE. (c) The densitometry analysis (of western blot) shows that p-tau (S202/T205) is significantly increased while the levels of PPP3CA and PPP3CB are reduced in CTE. Significantly different at *P<0.01. (d) The immunoreactivity of p-tau (Ser 396) is elevated while the immunoreactivity of PP3CA is reduced in CTE. Scale bar: white, 20 μm; black, 40 μm. (e) The densitometry analysis (of immunohistochemistry) shows that p-tau (S202/T205) is significantly increased, whereas the level of PPP3CA is reduced in CTE. Significantly different at **P<0.001.
Reduction of PPP3CA/PP2B activity is associated with tauopathy also in AD
Because the characteristics of CTE closely overlap with AD both clinically and pathologically, we examined whether PP2B activity is likely decreased in AD, which exhibits the most prevalent tauopathy. We conducted qPCR for PPP3CA in the AD subjects and confirmed downregulation, as shown in CTE subjects (Figure 4a). We also observed a similar inverse correlation between the PPP3CA levels and p-tau in AD, as shown in CTE (Figure 4b and c). As we expected, not only the immunoreactivity of PPP3CA was decreased but tau protein also was hyperphosphorylated (S199, S396, S205/T208) in AD (Figure 4d and e). Confocal microscopy showed that the immunoreactivity of PPP3CA was reduced but that the immunoreactivity of p-tau was elevated in AD (Figure 4f). Interestingly, we also observed the colocalization of PPP3CA with p-tau (S202/T205) in peripheral foci of neurofibrillary tangles in the cortex of AD.
Deregulation of PPP3CA expression is associated with the tauopathy in AD. (a) The mRNA levels of PPP3CA and PPP3CB are reduced in AD (n=8) compared to normal subjects (n=8). (b) The level of p-Tau (S202/T205) is elevated in AD, while the protein levels of PPP3CA and PPP3CB are reduced in AD. (c) The densitometry analysis shows that p-Tau (S202/T205) is significantly increased in AD, while the levels of PPP3CB are reduced in AD. *Significantly different at P<0.01. (d) The immunoreactivity of PPP3CA is reduced in AD, while the immunoreactivity of p-Tau (S396, S199 and S202/T205) is enhanced in AD. Scale bar: 50 μm (black); 20 μm (white). (e) The densitometry analysis (of immunohistochemistry) shows that p-tau (S202/T205) is significantly increased, whereas the level of PPP3CA is reduced in AD. Significantly different at **P<0.001. (f) The fluorescence intensity of PPP3CA is decreased in the temporal cortex of AD, while the fluorescence intensity of p-Tau (S202/T205) is increased. In part, the PPP3CA is colocalized with p-Tau (S202/T205) in the peripheral foci of the p-tau tangles detected in the temporal cortex of AD. The nuclei were counterstained with DAPI. Scale bar: 10 μm (white). The PPP3CA enzyme activity is significantly reduced in AD.
Inhibition of PPP3CA/PP2B activity leads to tau phosphorylation and tau aggregation
Following the confirmation of decreased PPP3CA activity and elevated tau phosphorylation in human CTE and AD samples, we examined whether PPP3CA inhibition would be sufficient to increase tau phosphorylation and aggregation using in vitro cell line models of tauopathy. To this end, we used Tau-BiFC cell lines (SH-SY5Y and HEK293), which enable the conventional fluorescence imaging of tau phosphorylation and aggregation (Figure 5a). Three different PP2A/PPP3CA inhibitors (okadaic acid, cyclosporine A and deltamethrin) increased the level of tau phosphorylation in both SH-SY5Y and HEK293 Tau-BiFC cell lines (Figure 5b). Okadaic acid and cyclosporine A treatment elevated the level of tau phosphorylation in a dose-dependent manner, whereas deltamethrin showed a declining trend after the threshold of 3 μM (Figure 5c). Similar results were observed in the HEK 293 cell line, with a higher intensity of tau phosphorylation due to okadaic acid (Figure 5d).
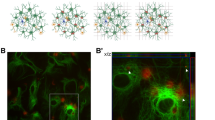
Protein phosphatase 2B (PP2B) (previous name of PPP3CA protein) inhibitors increase tau phosphorylation and tau aggregation in tau-BiFC cell lines. (a) Diagram of tau-BiFC maturation as an indication of tau aggregation. In tau-BiFC system, full-length tau is fused to non-fluorescent N- or C-terminal fragments of Venus fluorescence protein (VN173 or VC155), and both tau constructs were stably expressed in cells. Only when tau assembles, Venus protein could be matured, thereby activating its fluorescence signal. (b) PP2A/PP2B inhibitors induce tau aggregation in SH-SY5Y tau-BiFC cells. Cells were incubated with okadaic acid (OA; 30 nM), cyclosporine A (CsA; 30 μM) and deltamethrin (DM; 30 μM) for 29 h. Scale bar: 200 μm. (c) Quantification of BiFC-fluorescence increases in SH-SY5Y tau-BiFC cells at various concentrations of protein phosphatase (PP) inhibitors. (d) PP2A/PP2B inhibitors induce tau aggregation in HEK293 tau-BiFC cells. Cells were incubated with okadaic acid (OA; 30 nM), cyclosporine A (CsA; 30 μM) and deltamethrin (DM; 30 μM) for 29 h. Scale bar: 200 μm (white). (e) Quantification of BiFC-fluorescence increases in HEK293 tau-BiFC cells at various concentrations of PP inhibitors. Significantly different at *P<0.01; **P<0.001.
PPP3CA directly regulates tau phosphorylation
After confirming the involvement of PPP3CA catalytic activity in the dephosphorylation of tau, we further verified whether PPP3CA directly modulates tau phosphorylation (Figure 6). Because GSK3β is a well-known kinase of tau, we overexpressed GSK3β with PPP3CA and tau and determined the level of phosphorylated tau. Whereas the overexpression of GSK3β elevated tau phosphorylation at S202/T205 and S214, the co-transfection of PPP3CA reversed the effects of GSK3β and decreased the level of phosphorylated tau in a dose-dependent manner (Figure 6a). In addition, PPP3CB, another differentially expressed subunit of PPP3CA, reversed the effects of GSK3β and decreased the level of phosphorylated tau in a dose-dependent manner (Supplementary Figure 3). Regression analyses confirmed that the level of p-tau (S202/T205) was inversely correlated with the expression of PPP3CA (R=0.892; Figure 6b). In addition, we examined whether the catalytic subunit of PPP3CA is necessary and sufficient to phosphorylate tau (Figure 6c). To this end, we transfected cells with a catalytic site deletion mutant of PPP3CA (PPP3CAdeltaCAT) and verified that tau dephosphorylation did not occur with this mutant. The data showed that the catalytic activity of PPP3CA is indispensable for the dephosphorylation of p-tau. Moreover, we knocked down PPP3CA using shRNAs to demonstrate loss-of-function of PPP3CA (Figure 6d), resulting in a marked increase of tau phosphorylation in response to GSK3β.
PPP3CA regulates the dephosphorylation of tau. (a) PPP3CA reduces the level of p-tau (Ser214, Ser202/Thr205) by through GSK3β in a dose-dependent manner. (b) The level of phosphorylated tau is inversely correlated with the level of PPP3CA. (c) The catalytic site deletion mutant of PPP3CA (PPP3CAΔCat) did not reduce the level of p-tau (Ser202/Thr205). (d) The knockdown of PPP3CA/PPP3CA using shRNA increased the level of p-tau (Ser202/Thr205). (e) A schematic showing an in vivo animal model of AAV-shRNA injection. The AAV-shPPP3CA viral injection into the hippocampal region of mouse brain decreased the level of PPP3CA. Scale bar: 50 μm (white). (f) Knock down of PPP3CA increases the level of p-tau (S202/T205) in an animal model of TBI. The level of p-tau (Ser202/Thr205) was increased by AAV-shRNA for PPP3CA in the hippocampal region of mouse brain but not by AAV-shRNA control in response to TBI. (g) Densitometry analysis shows that the level of p-tau (Ser202/Thr205) is elevated in CA1, CA2, CA3 and DG (dentate gyrus) upon TBI. The level of p-tau was normalized to mCherry intensity. Significantly different at **P<0.001. (h) A schematic illustrating that the deregulation of protein phosphatases increases the level of p-tau and p-tau oligomer formation and leads to pathological neurofilament tangle formation in chronic traumatic encephalopathy (CTE).
PPP3CA/PP2B modulates tau dephosphorylation in in vivo animal model of CTE
To verify whether the downregulation of PPP3CA affects, the phosphorylation of tau in vivo, we transduced adeno-associated viral (AAV) vectors containing shRNA for PPP3CA into hippocampal regions of tau (P301L) transgenic mouse brains and measured the difference in tau phosphorylation compared with the control after multiple weight-drop-induced head injuries (Figure 6e). Consistent with the cellular model, the knockdown of PPP3CA using AAV-shPPP3CA markedly reduced the immunoreactivity of PPP3CA in the hippocampal regions of mice (Figure 6f). Furthermore, the immunoreactivity of p-tau (S202/T205) was robustly increased in the dentate gyrus after exposure to weight drop-induced head injuries (Figure 6g). The immunoreactivity of p-tau (S202/T205) was elevated in the region of PPP3CA knockdown using AAV-shPPP3CA, whereas p-tau (Ser202/Thr205) was not detected after AAV-shRNA control transduction (Figure 6h). Similarly, the immunoreactivity of p-tau (S199) was enhanced in hippocampal regions (CA1, CA3 and DG) with PPP3CA knockdown using AAV-shPPP3CA upon weight drop-induced multiple head injuries (Supplementary Figure 4).
Discussion
CTE is a progressive neurodegenerative disease that shows clinical symptoms including short-term memory loss, Parkinsonism, and gait and speech disabilities. Increasing evidence shows that a single or repetitive head injury is a risk factor for AD.27, 28 Importantly, CTE exhibits several neuropathological features of AD, such as tauopathy, neuropil neurites (NNs) and diffuse senile plaques. Indeed, the abnormal levels of hyperphosphorylated tau protein in neurons and astrocytes of CTE are similar or identical to those of neurofibrillary tangles in AD.29 However, although CTE neuropathologically resembles AD, the mechanism of how brain injury leads to neuropathological sequelae of tauopathy in CTE remains unknown.
In the present study, we analyzed the changes in the transcriptome associated with CTE. Although the pathophysiology of CTE is complex and involves broad changes in gene expression, reduced PPP3CA expression was observed in CTE. PPP3CA is a subunit of calcineurin (PP2B), which dephosphorylates tau, whose dysfunction has been implicated in the generation of both amyloid precursor protein and hyperphosphorylated tau, the two hallmarks of AD pathology.30 Previous studies have shown that the excitotoxin-induced dysfunction of serine/threonine PP activity induces tau phosphorylation in human neurons and that the downregulation of calcineurin (PPP3CA) causes tau hyperphosphorylation in AD patients and Huntington’s disease in mouse model brains.24, 31 Importantly, experimental systems showed that reduced PPP3CA expression likely leads to tau hyperphosphorylation in CTE, similar to AD.11 Notably, hyperphosphorylated tau was localized in neurofibrillary tangles and axonal arborizations, areas where the level of PPP3CA was inversely reduced in both CTE and AD cases. Moreover, in vitro experiments using phosphatase inhibitors on BiFC-Tau cells demonstrated the importance of serine/threonine PP activity for tau dephosphorylation.
Animal models of TBI have been developed over the last several decades. The weight-drop animal model of TBI mimics a repetitive brain injury in humans and provides several merits: (1) it is simple and easy to apply repetitive-hit consistently, and (2) it represents the pathophysiological features and symptoms of concussion.25 Thus, we established a weight-drop animal model of TBI and investigated how PPP3CA knockdown affects repetitive TBI-induced tau phosphorylation in MAPT (P301L) mice. Consistent with the cellular model, PPP3CA knockdown using AAV-shPPP3CA significantly increased the immunoreactivity of p-tau (S202/T205) in the dentate gyrus of P301L mice after exposure to repetitive head injuries. Together, these findings show that alterations in PP expression likely contribute to tauopathy in CTE, which suggests that modulating serine/threonine PP expression and activity could be a potential therapeutic strategy for preventing progressive tauopathy in CTE and other neurodegenerative diseases.
Accession codes
The sequencing data were deposited in the European Nucleotide Archive (ENA) database under accession no. ERP015139.
References
Thurman DJ, Branche CM, Sniezek JE . The epidemiology of sports-related traumatic brain injuries in the United States: recent developments. J Head Trauma Rehabil 1998; 13: 1–8.
Langlois JA, Rutland-Brown W, Wald MM . The epidemiology and impact of traumatic brain injury: a brief overview. J Head Trauma Rehabil 2006; 21: 375–378.
Nowinski C . Head Games: Football's Concussion Crisis from the NFL to Youth League. Drummond Publishing Group: East Bridgewater, MA, USA, 2006.
McKee AC, Cantu RC, Nowinski CJ, Hedley-Whyte ET, Gavett BE, Budson AE et al. Chronic traumaticencephalopathy in athletes: progressive tauopathy after repetitive head injury. J Neuropathol Exp Neurol 2009; 68: 709–735.
McKee AC, Stein TD, Kiernan PT, Alvarez VE . The neuropathology of chronic traumatic encephalopathy. Brain Pathol 2015; 25: 350–364.
DeKosky ST, Blennow K, Ikonomovic MD, Gandy S . Acute and chronic traumatic encephalopathies: pathogenesis and biomarkers. Nat Rev Neurol 2013; 9: 192–200.
Martland HAS . Punch drunk. J Am Med Assoc 1928; 91: 1103–1107.
Corsellis JA, Bruton CJ, Freeman-Browne D . The aftermath of boxing. Psychol Med 1973; 3: 270–303.
Stern RA, Daneshvar DH, Baugh CM, Seichepine DR, Montenigro PH, Riley DO et al. Clinical presentation of chronic traumatic encephalopathy. Neurology 2013; 81: 1122–1129.
Gandy S, Ikonomovic MD, Mitsis E, Elder G, Ahlers ST, Barth J et al. Chronic traumatic encephalopathy: clinical-biomarker correlations and current concepts in pathogenesis. Mol Neurodegener 2014; 9: 37.
McKee AC, Stern RA, Nowinski CJ, Stein TD, Alvarez VE, Daneshvar DH et al. The spectrum of disease in chronic traumatic encephalopathy. Brain 2013; 136: 43–64.
McKee AC, Cairns NJ, Dickson DW, Folkerth RD, Keene CD, Litvan I et al. The first NINDS/NIBIB consensus meeting to define neuropathological criteria for the diagnosis of chronic traumatic encephalopathy. Acta Neuropathol 2016; 131: 75–86.
Stiller JW, Weinberger DR . Boxing and chronic brain damage. Psychiatr Clin North Am 1985; 8: 339–356.
Mendez MF . The neuropsychological aspects of boxing. Int J Psychiatry Med 1995; 25: 249–262.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21.
Anders S, Pyl P, Huber W . HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169.
de Hoon MJ, Imoto S, Nolan J, Miyano S . Open source clustering software. Bioinformatics 2004; 20: 1453–1454.
Saldanha AJ . Java Treeview—extensible visualization of microarray data. Bioinformatics 2004; 20: 3246–3248.
Langfelder P, Horvath S . WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Tak H, Haque MM, Kim MJ, Lee JH, Baik JH, Kim Y et al. Himolecular fluorescence complementation; lighting-up Tau-Tau interaction in living cells. PLoS ONE 2013; 8: e81682.
Rahman A, Grundke-Iqbal I, Iqbal K . PP2B isolated from human brain preferentially dephosphorylates Ser-262 and Ser-396 of the Alzheimer disease abnormally hyperphosphorylated tau. J Neural Transm 2006; 113: 219–230.
Lee J, Hwang YJ, Shin JY, Lee WC, Wie J, Kim KY et al. Epigenetic regulation of cholinergicreceptor M1 (CHRM1) by histone H3K9me3 impairs Ca2+ signaling in Huntington’s disease. Acta Neuropathol 2013; 125: 727–739.
Rahman A, Ting K, Cullen KM, Braidy N, Brew BJ, Guillemin GJ . The excitotoxin quinolinic acid induces tau phosphorylation in human neurons. PLoS ONE 2009; 4: e6344.
Mychasiuk R, Farran A, Angoa-Perez M, Briggs D, Kuhn D, Esser MJ . A novel model of mild traumatic brain injury for juvenile rats. J Vis Exp 2014; 94: e51820.
Chiocco MJ, Zhu X, Walther D, Pletnikova O, Troncoso JC, Uhl GR et al. Fine mapping of Calcineurin(PPP3CA) gene reveals novel alternative splicing patterns, association of 5’UTR trinucleotide repeat with addiction vulnerability, and differential isoform expression in Alzheimer’s disease. Subst Use Misuse 2010; 45: 1809–1826.
Corsellis JA, Brierley JB . Observations on the pathology of insidious dementia following head injury. J Ment Sci 1959; 105: 714–720.
Rudelli R, Strom JO, Welch PT, Ambler MW . Posttraumatic premature Alzheimer’s disease. Neuropathologic findings and pathogenetic considerations. Arch Neurol 1982; 39: 570–575.
Schmidt ML, Zhukareva V, Newell KL, Lee VM, Trojanowski JQ . Tau isoform profile and phosphorylation state in dementia pugilistica recapitulate Alzheimer’s disease. Acta Neuropathol 2001; 101: 518–524.
Braithwaite SP, Stock JB, Lombroso PJ, Nairn AC . Protein phosphatases and Alzheimer's disease. Prog Mol Biol Transl Sci 2012; 106: 343–379.
Gratuze M, Noël A, Julien C, Cisbani G, Milot-Rousseau P, Morin F et al. Tau hyperphosphorylation and deregulation of calcineurin in mouse models of Huntington's disease. Hum Mol Genet 2015; 24: 86–99.
Acknowledgements
We thank Thomas Bleazard at Faculty of Medical and Human Sciences, University of Manchester for discussion and assistance with manuscript preparation. This work was financially supported by grants from Macrogen Inc. (grant no. MGR14-02), NIH R01 NS067283 (HR), Brain Science Flagship Grant (2E26200 and 2N41600 HR) and an intramural grant (2E26640 CHK) from the Korea Institute of Science and Technology. The authors gratefully acknowledge the use of the resources and facilities at the Edith Nourse Rogers Memorial Veterans Hospital (Bedford, MA, USA). This work was financially supported by grants from the National Institute of Neurological Disorders and Stroke (1U01NS086659-01, Department of Veterans Affairs, the Veterans Affairs Biorepository (CSP 501), the National Institute of Aging Boston University Alzheimer’s Disease Center (P30AG13846; supplement 0572063345–5), the National Institute of Aging Boston University Framingham Heart Study (R01AG1649), and the Concussion Legacy Foundation. This work was also financially supported by unrestricted gifts from the Andlinger Foundation, the World Wrestling Entertainment and the National Football League.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Seo, JS., Lee, S., Shin, JY. et al. Transcriptome analyses of chronic traumatic encephalopathy show alterations in protein phosphatase expression associated with tauopathy. Exp Mol Med 49, e333 (2017). https://doi.org/10.1038/emm.2017.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2017.56
This article is cited by
-
Lipocalin-2 Is a Key Regulator of Neuroinflammation in Secondary Traumatic and Ischemic Brain Injury
Neurotherapeutics (2023)
-
Neuropathology in chronic traumatic encephalopathy: a systematic review of comparative post-mortem histology literature
Acta Neuropathologica Communications (2022)
-
The role of protein phosphatase 2A tau axis in traumatic brain injury therapy
Beni-Suef University Journal of Basic and Applied Sciences (2022)
-
Evidence of traumatic brain injury in headbutting bovids
Acta Neuropathologica (2022)
-
Mouse closed head traumatic brain injury replicates the histological tau pathology pattern of human disease: characterization of a novel model and systematic review of the literature
Acta Neuropathologica Communications (2021)