Abstract
Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) is a novel strategy for cancer therapy, but only two inhibitors of NAMPT (FK866 and CHS828) have progressed into clinical trials. This study seeks to compare a novel potent NAMPT inhibitor, MS0, with a classical inhibitor FK866 in their biological activity and molecular binding mode, thereby contributing to future chemical optimization and a further understanding of the action mode of NAMPT inhibitors. The IC50 values of MS0 and FK866 in inhibition of recombinant human NAMPT activity were 9.08±0.90 and 1.60±0.32 nmol/L, respectively. Consistently, FK866 exerted better antiproliferation in 6 human cancer cell lines (HepG2, A2780, 95-D, A549, U2OS and U266) than MS0 with IC50 values nearly 12-fold to 225-fold lower than those of MS0. Co-crystal structures of wild-type human NAMPT complexed with MS0 or FK866 were elucidated, which revealed that MS0 did not interact with Ser241. The hydrogen bond mediated by crystallographic water between MS0 and His191 or Val350 of NAMPT did not exist in FK866. Instead, FK866 exhibited hydrophobic interactions with Arg349. Based on the activity assays and crystal structure analyses, we elaborate the reason why the antiproliferation activity of MS0 was not as good as that of FK866, which would contributes to the current understanding of the mode of action of NAMPT inhibitors and will also contribute to further development of anticancer drugs in the future.
Similar content being viewed by others
Introduction
Nicotinamide phosphoribosyltransferase (NAMPT) is the rate-limiting enzyme in the NAD biosynthesis pathway in mammals, which regulates total cellular NAD levels and mitochondrial NAD levels1,2,3. There are three names for the enzyme of interest: NAMPT, PBEF, and Visfatin4,5,6. In 1994, NAMPT was first found in peripheral blood lymphocytes, which can promote proliferation of pre-B cells7. In 2005, it was reported as an adipokine called Visfatin. NAMPT has many biological functions, and one of the most important functions is maintaining an energy supply by synthesizing NAD, which plays a critical role in cell survival. Owing to its importance in cellular pathophysiological processes and its close relationship with the occurrence of and development of tumors, NAMPT is a promising target for the discovery of novel antitumor agents8,9.
Our team has conducted many studies on this enzyme, particularly regarding the inhibition of cancer cells. We have discovered a series of effective NAMPT inhibitors in our previous work10. Meanwhile, other compounds have been reported during these years and have also shown pharmacological activities11,12,13,14,15. However, there are only 2 main inhibitors of NAMPT that have progressed into clinical trials. FK866 has progressed to phase II clinical trials, but it has low bioavailability and rapid intravenous clearance16,17. CHS828 has progressed to phase I preclinical trials, but it has dose-limiting side effects such as thrombocytopenia18,19. Thus, much work remains to be done in this area. This article is focused on the level of the crystal-structure to compare our most effective compound, MS0, with the classical NAMPT inhibitor, FK866, to explain the difference in activity between these two compounds.
Materials and methods
Plasmid construction and protein expression
The cDNA sequence of human NAMPT was amplified by PCR from a pGex-6p-3-hNAMPT plasmid (kindly gifted from Dr Shui-qing YE, University of Missouri). The PCR products were digested and cloned into a pET21a+ vector using NdeI and XhoI restriction enzymes. A point mutation was introduced by the quick change site-directed mutagenesis method using the constructed pET21a+-hNAMPT plasmid as a template. Primer pairs are listed in the Supplemental Materials and Methods. All mutations were validated by DNA sequencing. His-tagged wild-type NAMPT, NAMPT mutants and NMNAT1 were expressed and purified by our previous methods20,21, also detailed in the Supplemental Materials and Methods.
Determination of IC50 values for NAMPT inhibitors
Determination of the IC50 for NAMPT inhibitors was carried out using recombinant human NAMPT according to our previously reported method21, which is based on measuring the fluorescence of a nicotinamide mononucleotide (NMN) derivative that results from the enzymatic production of NMN (see the Supplemental Materials and Methods).
Protein crystallization
Crystals of wild-type human NAMPT complexed with MS0 were obtained at 295 K by the hanging-drop vapor-diffusion method in 24-well plates (Hampton Research Inc, CA, USA). Each well was filled with 500 μL of reservoir solution. Drops consisting of 1 μL NAMPT solution and 1 μL reservoir solution were placed onto 22 mm silicone circular cover slides. The protein (20 mg/mL) was incubated with 1 mmol/L MS0 at 277 K for 30 min before the crystallization setup. The reservoir solution contained 50 mmol/L phosphate buffer (pH 9.2), 28% (w/v) PEG-3350, 180 mmol/L NaCl and 5 mmol/L DTT. There were two copies of the NAMPT-MS0 complex in each asymmetric unit.
Data collection and processing
X-ray diffraction data were collected on a CCD at the BL-18U beamline at Shanghai Synchrotron Radiation Facility. Data on the free enzyme crystal were collected at 1.46 Å resolution and 100 K, and native diffraction data sets were collected on the other crystals. The diffraction images were processed and scaled with the XIA2. The data processing statistics are summarized in Extended Data Table 1.
Structure determination and refinement
The atomic model of NAMPT was built with Phoenix. After one cycle of refinement at 1.46 Å resolution with Phoenix, a model for the dimer of NAMPT was used to solve the structure of the human NAMPT-MS0 complex by molecular replacement. Refinement of these structures was carried out using Phoenix and Coot. The refinement statistics are summarized in Extended Data Table 1.
Protein stability assays
Protein thermostability was tested by a microscale fluorescent thermal stability assay using SYPRO orange protein gel stain (Sigma-Aldrich, USA), which reacts with native hydrophobic regions embedded in the protein interior and acts as a sensor for the overall integrity of the folded state. Stock SYPRO orange dye was diluted (1:1000) with 50 mmol/L phosphate buffer (pH 9.2), 180 mmol/L NaCl and 5 mmol/L DTT. The protein to be tested (∼5 μg) was diluted in the same buffer to a final volume of 33 μL. One μL of the diluted dye was added and thoroughly mixed with the protein. The reaction mixture was incubated at room temperature for 15 min, subsequently transferred to a sub-micro quartz fluorometer cuvette (Starna Cells, Inc, Atascadero, CA, USA) and heated in a controlled environment with a ramp rate of 1 °C/min over the temperature range of 25–90 °C, using a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies, USA) to detect the dye. The excitation wavelength was 490 nm, while the emission wavelength was 575 nm.
Results
Inhibitory effect of MS0 and FK866 on human cancer cell lines
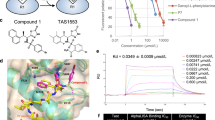
Through our previous work, we discovered a novel structure compound, MS0 (Figure 1A), that is a highly effective NAMPT inhibitor. FK866 (Figure 1A) is a classic NAMPT inhibitor with high inhibition efficacy. First, we evaluated the enzyme inhibition activity of these two compounds using our previously established method21. The results showed that the IC50 value of MS0 was nearly 6 times higher than that of FK866 (9.08±0.90 vs 1.60±0.32 nmol/L) (Figure 1B and 1C). Furthermore, we examined their effects on the cell viability of a hepatocellular carcinoma cell line, HepG2, using the cell counting kit-8 (CCK-8) method22. The difference between the IC50 value of MS0 (IC50=510.01±162.09 nmol/L) and FK866 (IC50=2.21±0.21 nmol/L) is nearly 250-fold (Figure 1D and 1E). To evaluate cell viability effects more extensively, we tested both inhibitors using the sulforhodamine B protein staining (SRB) method23 on 6 human cancer cell lines, including ovarian cancer cell line A2780, metastatic lung cancer cell line 95-D, lung adenocarcinoma cell line A549, osteosarcoma cell line U2OS, and multiple myeloma cell line U266 and HepG2. As shown in Figure 1F, FK866 has better antiproliferation activity on all 6 human cancer cell lines with IC50 values nearly 12-fold to 225-fold lower than those of MS0.
Activity and structure differences between MS0 and FK866. (A) Structural differences between MS0 and FK866. (B) Inhibitory potency (IC50) of MS0 on NAMPT activity. (C) Inhibitory potency (IC50) of FK866 on NAMPT activity. (D) Concentration response curve of MS0 on HepG2 cells after 48 h of treatment. (E) Concentration response curve of FK866 on HepG2 cells after 48 h of treatment. (F) IC50 of MS0 and FK866 inhibition on various human cancer cell lines (SRB assay after 72 h of treatment). Data are expressed as the mean±SEM.
Nu-PAGE and Coomassie Blue staining analysis of purified human NAMPT
As Figure 2A shown, both NAMPT and its complex NAMPT+MS0 have two bands in the Nu-PAGE, indicating that purified NAMPT would exist in dimer form.
Protein stability and crystals of NAMPT with and without MS0. (A) Protein purification of NAMPT; NAMPT stands for NAMPT without MS0, and NAMPT+MS0 stands for NAMPT with MS0. (B) CPM ramping assay of NAMPT, temperature range of 45–60 °C. (C) Crystals of NAMPT with and without MS0.
Protein stability assays
In this study, we performed protein stability assays to further verify that MS0 can interact with NAMPT. We used the cysteine-reactive fluorescent dye CPM to test if MS0 can bind to NAMPT. According to the results shown in Figure 2B and 2C, the CPM ramping protein assay showed that the NAMPT-MS0 complex (green trace) has a higher melting temperature (Tm), which indicates better protein thermostability compared to NAMPT alone.
Crystallization of NAMPT
We determined the crystal structure of NAMPT with and without MS0 at 1.46 Å and 2.00 Å resolutions, respectively, to elucidate the nature of the enzymatic active site, as well as the catalytic mechanism (Figure 3). The two active sites of NAMPT lie at the dimer interface, where the substrates and products bind. The pyridine moiety of MS0 lies between Phe193 from one chain of NAMPT and Tyr18' from the other, showing π-stacking interactions (Figure 3A). Crystallographic water was observed mediating a hydrogen bond network among the thiourea nitrogens, the backbone carbonyl of Val242, and the Asp219 side chains (Figure 3B). The carbonyl oxygen of the amide group of MS0 is pointed perpendicularly toward the side chain of Arg311 of NAMPT. Hydrophobic contacts were noted between the MS0 piperidine carbon atom and the side chains of Ile309 and Pro273 of NAMPT (Figure 3C).
Crystal structure analysis of NAMPT with MS0 and FK866. (A–C) Crystal structure analysis of NAMPT with MS0. MS0 and the two residues of the protein are labeled. MS0 atoms are shown in yellow. The bound position of MS0 is also shown in red and cyan. (A) π-stacking interactions with NAMPT and the head group of MS0. (B) Hydrogen bond between NAMPT and the middle group of MS0. (C) Hydrophobic interactions and hydrogen bond between NAMPT and the tail group of MS0. (D–F) Crystal structure analysis of NAMPT with FK866. FK866 and the two residues of the protein are labeled. FK866 atoms are shown in orange. The bound position of FK866 is also shown in green and orange. (D) π-stacking interactions with NAMPT and the head group of FK866. (E) Hydrogen bond between NAMPT and the middle group of FK866. (F) Hydrophobic bond between NAMPT and the tail group of FK866.
Structure variation between NAMPT with MS0 and NAMPT with FK866
As shown in Figure 1A, MS0 has a very similar molecular structure to FK866 that can be divided into 3 parts. Obviously, MS0 lacks part IV, which may prompt FK866 to be more active than MS0. However, further study on the crystal structures still needs to be performed.
To determine the activity difference between MS0 and FK866 at the molecular level, we compared the co-crystal structure of NAMPT with MS0 and with FK866. According to the results, both co-crystal structures have π-stacking interactions in the pyridine moiety between Phe193 and Tyr18' (Figure 3A and 3D). As Figure 3E shown, a hydrogen bond was mediated by crystallographic water between the amide nitrogen of FK866 and Asp219, Ser241 and Val242 of NAMPT. Furthermore, MS0 does not interact with Ser241 of NAMPT, which may make FK866 more stable than MS0 in binding to NAMPT (Figure 3B). FK866 also forms a hydrogen bond between Ser275 and the amide oxygen, whereas this hydrogen bond cannot be observed with MS0 (Figure 3B and 3E). These variations may lead to the activity difference between FK866 and MS0. In addition, the hydrogen bond mediated by crystallographic water between MS0 and His191 and Val350 would disappear in FK866, since His191 and Val 350 do not exist near parts III and IV of this compound; instead, this area of FK866 exhibits hydrophobic interactions with Arg349 (Figure 3C and 3F). Thus, these changes may contribute to the activity difference between FK866 and MS0.
Discussion
FK866 is a classic NAMPT inhibitor with high efficacy. Comparative studies of FK866 and other compounds can provide a new view for exploring future NAMPT inhibitors. MS0 is our previously discovered potent NAMPT inhibitor with novel structure. Thus, we chose MS0 as the compound for comparison with FK866 to find more details about inhibitor interactions with NAMPT for future chemical discovery.
Developing active compounds to target NAMPT is important and promising because NAMPT plays significant roles in a variety of biological events and has drawn much attention in several different fields, including NAD biology, metabolism, and immune response24,25. Ample inhibitors11,12 targeting NAMPT have been developed in recent years since FK86616 and GMX1778 (previously known as CHS828)26 were initially discovered; however, no compound has been confirmed as a targeted drug because of the lack of understanding about the NAMPT action mode. Recently, NAMPT-catalyzed phosphoribosylation27 has been reported as a relevant biological mechanism of action. Specifically, inhibitors decrease the enzyme activity through NAMPT-catalyzed phosphoribosylation to produce pRib adducts. Based on this mode, we can explain the phenomenon that biochemically potent NAMPT inhibitors do not always exhibit favorable effects in cell culture assessments, even when the inhibitors have the same permeability. For instance, GNE-61728 and GMX177829 can inhibit NAMPT activity significantly and were efficacious when tested in a variety of human cancer cell lines. GNE-64330 is a close structural analogue of GNE-617, but it showed relatively weaker antiproliferative properties than GNE-617 and GMX1778. GNE-200 and GNE-201 are close analogues of GNE-617 and GNE-643, respectively, but they lack the nitrogen atoms required for pRib adduct formation. Therefore, GNE-200 and GNE-201 showed the worst efficacy compared to GNE-617 and GMX1778 (Figure 4). By using the NAMPT-catalyzed phosphoribosylation theory, we can divide these 5 compounds into 3 classes: A) those that can form NAMPT-dependent pRib adducts and bind to the enzyme with high affinity (GNE-617 and GMX1778), B) those that can form NAMPT-dependent pRib adducts but do not retain potent binding affinity (GNE-643), 3) those that cannot form NAMPT-dependent pRib adducts (GNE-200 and GNE-201) (Figure 4). These diverse biochemical profiles are tightly correlated with the inhibitory potencies of the compounds in cells, while those belonging to classes A, B, and C exhibit strong, medium, and weak antiproliferative properties, respectively. Although such trends still need further verification on additional NAMPT inhibitors to ultimately confirm their usefulness, this comparison suggests that future designs for cellularly active NAMPT inhibitors should target the class A profile (Figure 4). However, MS0 and FK866 do not belong to these categories.
Compound classifications. Class A contains GMX1778, GNE-617 and pRib-GNE-617; Class B contains GNE-643; Class C contains GNE-200 and GNE-201.
We found that MS0 is truly less active than FK866 in vitro, probably due to the variation in molecular structures (Figure 1A). We find that MS0 is shorter than FK866 after being divided into 4 parts, of which part IV most likely to influence their activities. Therefore, we performed crystal structure studies to further elucidate the activity differences. Comparing the co-crystal structure of NAMPT complexed with MS0 to that of NAMPT with FK866, we found that both structures have the same interactions in the pyridine moiety of part I, forming π-stacking interactions, which had been shown to be the key mode of action in other studies. In addition to the π-stacking interactions, hydrogen bond is another intermolecular force that was observed. We observe that, in part I of the molecular structures, the hydrogen bond of FK866 is much stronger than that of MS0, which could be the reason for different activities between the inhibitors. The tail parts of the compounds are different, as well. FK866 exhibits hydrophobic binding with Arg349 of NAMPT compared to the hydrogen bonds mediated by crystallographic water between MS0 and His191 and Val 350 of NAMPT. These differences may allow FK866 more flexibility to fit into the binding site.
Above all, for future NAMPT inhibitor development, newly designed compounds are recommended to possess the following characteristics for better efficacy: preservation of the pyridine residue, more hydrogen bonding in part I of the molecule, a longer structure in part IV and more hydrogen bonding in part IV.
Author contribution
Chao-yu MIAO and Qiang ZHAO designed the research and revised the manuscript; Sai-long ZHANG and Zhen-lin YANG performed the protein crystallization study. Shuo HAN contributed to data collection and the refinement statistics. Sai-long ZHANG and Tian-ying XU performed the other experiments, analyzed the data and wrote the manuscript.
References
Wang P, Miao CY . NAMPT as a therapeutic target against stroke. Trends Pharmacol Sci 2015; 36: 891–905.
Li ZY, Song J, Zheng SL, Fan MB, Guan YF, Qu Y, et al. Adipocyte Metrnl antagonizes insulin resistance through PPARgamma signaling. Diabetes 2015; 64: 4011–22.
Zheng SL, Li ZY, Song J, Liu JM, Miao CY . Metrnl: a secreted protein with new emerging functions. Acta Pharmacol Sin 2016; 37: 571–9.
Preiss J, Handler P . Enzymatic synthesis of nicotinamide mononucleotide. J Biol Chem 1957; 225: 759–70.
Martin PR, Shea RJ, Mulks MH . Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J Bacteriol 2001; 183: 1168–74.
Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol 2002; 32: 3225–34.
Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I . Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol 1994; 14: 1431–7.
Jieyu H, Chao T, Mengjun L, Shalong W, Xiaomei G, Jianfeng L, et al. Nampt/Visfatin/PBEF: a functionally multi-faceted protein with a pivotal role in malignant tumors. Curr Pharm Des 2012; 18: 6123–32.
Buldak RJ, Buldak L, Polaniak R, Kukla M, Birkner E, Kubina R, et al. Visfatin affects redox adaptative responses and proliferation in Me45 human malignant melanoma cells: an in vitro study. Oncol Rep 2013; 29: 771–8.
Xu TY, Zhang SL, Dong GQ, Liu XZ, Wang X, Lv XQ, et al. Discovery and characterization of novel small-molecule inhibitors targeting nicotinamide phosphoribosyltransferase. Sci Rep 2015; 5: 10043.
Christensen MK, Erichsen KD, Olesen UH, Tjornelund J, Fristrup P, Thougaard A, et al. Nicotinamide phosphoribosyltransferase inhibitors, design, preparation, and structure-activity relationship. J Med Chem 2013; 56: 9071–88.
Galli U, Travelli C, Massarotti A, Fakhfouri G, Rahimian R, Tron GC, et al. Medicinal chemistry of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors. J Med Chem 2013; 56: 6279–96.
Matheny CJ, Wei MC, Bassik MC, Donnelly AJ, Kampmann M, Iwasaki M, et al. Next-generation NAMPT inhibitors identified by sequential high-throughput phenotypic chemical and functional genomic screens. Chem Biol 2013; 20: 1352–63.
Zheng X, Bauer P, Baumeister T, Buckmelter AJ, Caligiuri M, Clodfelter KH, et al. Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (Nampt) inhibitors. J Med Chem 2013; 56: 4921–37.
Giannetti AM, Zheng X, Skelton NJ, Wang W, Bravo BJ, Bair KW, et al. Fragment-based identification of amides derived from trans-2-(pyridin-3-yl)cyclopropanecarboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT). J Med Chem 2014; 57: 770–92.
Hasmann M, Schemainda I . FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res 2003; 63: 7436–42.
Holen K, Saltz LB, Hollywood E, Burk K, Hanauske AR . The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Invest New Drugs 2008; 26: 45–51.
Ravaud A, Cerny T, Terret C, Wanders J, Bui BN, Hess D, et al. Phase I study and pharmacokinetic of CHS-828, a guanidino-containing compound, administered orally as a single dose every 3 weeks in solid tumours: an ECSG/EORTC study. Eur J Cancer 2005; 41: 702–7.
von Heideman A, Berglund A, Larsson R, Nygren P . Safety and efficacy of NAD depleting cancer drugs: results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother Pharmacol 2010; 65: 1165–72.
Lv XQ, Zhang RY, Xu XW, Guan YF, Miao CY . Expression, purification, and enzymatic activity assay of nicotinamide mononucleotide adenylyltransferase. Acad J Sec Mil Med Univ 2010; 31: 1251–4.
Zhang RY, Qin Y, Lv XQ, Wang P, Xu TY, Zhang L, et al. A fluorometric assay for high-throughput screening targeting nicotinamide phosphoribosyltransferase. Anal Biochem 2011; 412: 18–25.
Liu Y, Qi Y, Bai ZH, Ni CX, Ren QH, Xu WH, et al. A novel matrine derivate inhibits differentiated human hepatoma cells and hepatic cancer stem-like cells by suppressing PI3K/AKT signaling pathways. Acta Pharmacol Sin 2017; 38: 120–32.
Wang YQ, Shen AJ, Sun JY, Wang X, Liu HC, Zhang MM, et al. Targeting Hsp90 with FS-108 circumvents gefitinib resistance in EGFR mutant non-small cell lung cancer cells. Acta Pharmacol Sin 2016; 37: 1587–96.
Garten A, Petzold S, Korner A, Imai S, Kiess W . Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol Metab 2009; 20: 130–8.
Imai S . Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr Pharm Des 2009; 15: 20–8.
Hjarnaa PJ, Jonsson E, Latini S, Dhar S, Larsson R, Bramm E, et al. CHS 828, a novel pyridyl cyanoguanidine with potent antitumor activity in vitro and in vivo. Cancer Res 1999; 59: 5751–7.
Takahashi R, Nakamura S, Yoshida T, Kobayashi Y, Ohkubo T . Crystallization of human nicotinamide phosphoribosyltransferase. Acta Crystallogr Sect F Struct Biol Cryst Commun 2007; 63: 375–7.
Zheng X, Bair KW, Bauer P, Baumeister T, Bowman KK, Buckmelter AJ, et al. Identification of amides derived from 1H–pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT). Bioorg Med Chem Lett 2013; 23: 5488–97.
Watson M, Roulston A, Belec L, Billot X, Marcellus R, Bedard D, et al. The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol Cell Biol 2009; 29: 5872–88.
Oh A, Ho YC, Zak M, Liu Y, Chen X, Yuen PW, et al. Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase. Chembiochem 2014; 15: 1121–30.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No 81130061 and 81373414 to Chao-yu MIAO) and the Shanghai Municipal Science and Technology Project (No 16431901400 to Chao-yu MIAO and No 16140904500 to Tian-ying XU).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary information is available at the website of Acta Pharmacologica Sinica.
Supplementary information
Supplemental Data
Supplemental Materials and Methods (DOC 80 kb)
Rights and permissions
About this article
Cite this article
Zhang, Sl., Xu, Ty., Yang, ZL. et al. Crystal structure-based comparison of two NAMPT inhibitors. Acta Pharmacol Sin 39, 294–301 (2018). https://doi.org/10.1038/aps.2017.80
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2017.80
Keywords
This article is cited by
-
Structural insights into Xanthomonas campestris pv. campestris NAD+ biosynthesis via the NAM salvage pathway
Communications Biology (2024)
-
Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) with OT-82 induces DNA damage, cell death, and suppression of tumor growth in preclinical models of Ewing sarcoma
Oncogenesis (2020)