Abstract
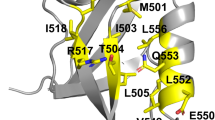
EXPERIMENTAL1–6 and simulation7 studies show that small mono-meric proteins fold in one kinetic step, which entails overcoming the free-energy barrier between the unfolded and the native protein through a transition state8,9. Two models of transition state formation have been proposed: a 'nonspecific' one in which it depends on the formation of a sufficient number of native-like contacts regardless of what amino acids are involved10–12, and a 'specific' one, in which it depends on formation of a specific subset of the native structure (a folding nucleus)8,13,14. The latter requires that some amino acids form most of their contacts in the transition state, whereas others only do so on reaching the native conformation. If so, mutations affecting the stability of the transition state nucleus should have a greater effect on the folding kinetics than mutations elsewhere, and the residues involved should be evolutionarily conserved. Lattice-model simulations and experiments8,13–16 suggest that such mutations exist. Here we present a method for determining the folding nucleus of a protein with known structure with two-state folding kinetics. This method is based on the alignment of many sequences designed to fold into the native conformation of a protein to identify the positions where amino acids are most conserved in designed sequences. The method is applied to chymotrypsin inhibitor 2 (CI2), a protein whose transition state has been previously studied by protein engineering14–16. The involvement of residues in folding nucleus of CI2 is clearly correlated with their conservation in design, and the residues forming the nucleus are highly conserved in 23 natural sequences homologous to CI2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Jackson, S. E. & Fersht, A. R. Biochemistry 30, 10428–10435 (1991).
Alexander, P., Orban, J. & Bryan, P. Biochemistry 31, 7243–7248 (1992).
Schindler, T., Herrler, M., Marahiel, M. & Schmid, F. X. Nature struct. Biol. 2, 663–673 (1995).
Sosnick, T. R., Mayne, L., Hiller, R. & Englander, S. W. Nature struct. Biology 1, 149–156 (1994).
Viguera, A. R., Martinez, J. C., Filiminov, V. V., Mateo, P. L. & Serrano, L. Biochemistry 33, 2142–2150 (1994).
Kragelund, B. B., Robinson, C. V., Knudsen, J. & Dobson, C. M. Biochemistry 34, 7117–7124 (1995).
Gutin, A. M., Abkevich, V. I. & Shakhnovich, E. I. Biochemistry 34, 3066–3076 (1995).
Abkevich, V. I., Gutin, A. M. & Shakhnovich, E. I. Biochemistry 33, 10026–10036 (1994).
Guo, Z. & Thirumalai, D. Biopolymers 35, 137–139 (1995).
Shakhnovich, E. I., Farztdinov, G., Gutin, A. M. & Karplus, M. Phys. Rev. Lett. 67, 1665–1668 (1991).
Sali, A., Shakhnovich, E. I. & Karplus, M. Nature 369, 248–251 (1994).
Wolynes, P. G., Onuchic, J. N. & Thiramulai, D. Science 267, 1619–1620 (1995).
Fersht, A. Proc. natn. Acad. Sci. U.S.A 92, 10869–10873 (1995).
Itzhaki, L., Oltzen, D. & Fersht, A. J. molec. Biol. 254, 260–288 (1995).
Jackson, S. E., elMasry, N. & Fersht, A. Biochemistry 32, 11270–11278 (1993).
Oltsen, D. E., Itzhaki, L., elMasry, N., Jackson, S. E. & Fersht, A. Proc. natn. Acad. Sci. U.S.A. 91, 10422–10425 (1994).
Shakhnovich, E. I. Phys. Rev. Lett. 72, 3907–3910 (1994).
Sander, C. & Schneider, R. Proteins 9 56–68 (1991).
Myazawa, S. & Jernigan, R. Macromolecules 18, 534–552 (1985).
Kolinski, A., Godzik, A. & Skolnick, J. J. chem. Phys. 98, 7420–7433 (1993).
Shakhnovich, E. I. & Gutin, A. M. Proc. natn. Acad. Sci. U.S.A. 90, 7195–7199 (1993).
Shakhnovich, E. I. & Gutin, A. M. Prot. Engng 6, 793–800 (1993).
Bowie, J. U., Luthy, R. & Eisenberg, D. Science 253, 164–169 (1991).
Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N. & Teller, E. J. chem. Phys 21, 1087–1092 (1953).
Abkevich, V. I., Gutin, A. M. & Shakhnovich, E. I. J. chem. Phys 101, 6052–6062 (1994).
Goldstein, R., Luthey-Schultzen, Z. A. & Wolynes, P. G. Proc. natn. Acad. Sci. U.S.A. 89, 4918–4922 (1992).
Sali, A., Shakhnovich, E. I. & Karplus, M. J. molec. Biol. 235, 1614–1636 (1994).
Hao, M.-H. & Scheraga, H. J. ehem. Phys 102, 1334–1339 (1995).
Gutin, A. M., Abkevich, V. I. & Shakhnovich, E. I. Proc. natn. Acad. Sci. U.S.A. 92, 1282–1286 (1995).
Matouschek, A., Kellis, J., Serrano, L., Bycroft, M. & Fersht, A. Nature 346, 440–445 (1990).
Matouschek, A., Serrano, L. & Fersht, A. J. molec. Biol. 224, 819–835 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Shakhnovich, E., Abkevich, V. & Ptitsyn, O. Conserved residues and the mechanism of protein folding. Nature 379, 96–98 (1996). https://doi.org/10.1038/379096a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/379096a0
This article is cited by
-
Kinetic and thermodynamic studies reveal chemokine homologues CC11 and CC24 with an almost identical tertiary structure have different folding pathways
BMC Biophysics (2017)
-
Understanding the Molecular Dynamics of Type-2 Diabetes Drug Target DPP-4 and its Interaction with Sitagliptin and Inhibitor Diprotin-A
Cell Biochemistry and Biophysics (2014)
-
Fast Side Chain Replacement in Proteins Using a Coarse-Grained Approach for Evaluating the Effects of Mutation During Evolution
Journal of Molecular Evolution (2011)
-
Structural and functional constraints in the evolution of protein families
Nature Reviews Molecular Cell Biology (2009)
-
Robustness of the residue conservation score reflecting both frequencies and physicochemistries
Amino Acids (2008)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.