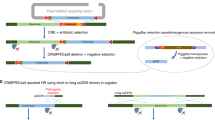
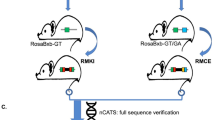
Key Points
-
Genetic engineering has traditionally been done in Escherichia coli using restriction enzymes to cleave DNA, and using DNA ligases to join them. However, there are several limitations to this approach, especially when large DNA molecules require engineering, because even rare restriction enzymes occur over large stretches of DNA. The generation of transgenic and mouse knockout constructs in E. coli is also hampered by the difficulty of finding appropriately placed restriction-enzyme cleavage sites.
-
Genetic engineering in yeast alleviates these problems because it relies on homologous recombination rather than on restriction enzymes and DNA ligases to generate recombinant DNA molecules.
-
A principal limitation of genetic engineering in yeast is that yeast artificial cloning vectors (YACs), developed for cloning large DNA molecules, are often unstable, and YAC transgenic mice are difficult to make. As a result, more stable vectors, such as bacterial artificial chromosomes (BACs), are often used instead.
-
Phage-based E. coli recombination systems have been developed that now allow large DNA molecules cloned into BACs to be modified by homologous recombination, similarly to what occurs in yeast. These E. coli recombination systems have many of the advantages of yeast recombination, but few of its disadvantages.
-
This new form of chromosome engineering, termed recombineering, makes it possible to introduce virtually any type of mutation into a BAC using PCR-amplified, linear, double-stranded DNA targeting cassettes that have short regions of homology at their ends, or single-stranded oligonucleotides.
-
Recombineering greatly decreases the time it takes to create transgenic mouse models by conventional means. It also facilitates many kinds of genomic experiment that have otherwise been difficult to carry out, and it should enhance functional genomic studies by providing better mouse models and a more refined genetic analysis of the mouse genome.
Abstract
Highly efficient phage-based Escherichia coli homologous recombination systems have recently been developed that enable genomic DNA in bacterial artificial chromosomes to be modified and subcloned, without the need for restriction enzymes or DNA ligases. This new form of chromosome engineering, termed recombinogenic engineering or recombineering, is efficient and greatly decreases the time it takes to create transgenic mouse models by traditional means. Recombineering also facilitates many kinds of genomic experiment that have otherwise been difficult to carry out, and should enhance functional genomic studies by providing better mouse models and a more refined genetic analysis of the mouse genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Clarke, A. R. Manipulating the germline: its impact on the study of carcinogenesis. Carcinogenesis 21, 435–441 (2000).
Gao, X., Kemper, A. & Popko, B. Advanced transgenic and gene-targeting approaches. Neurochem. Res. 24, 1181–1188 (1999).
Macleod, K. F. & Jacks, T. Insights into cancer from transgenic mouse models. J. Pathol. 187, 43–60 (1999).
Lobe, C. G. & Nagy, A. Conditional genome alteration in mice. Bioessays 20, 200–208 (1998).
Murphy, K. C. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180, 2063–2071 (1998).The first use of phage-recombination functions for generating recombinant molecules using linear double-stranded DNA.
Zhang, Y., Buchholz, F., Muyrers, J. P. & Stewart, A. F. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet. 20, 123–128 (1998).The first demonstration of recombineering using PCR-amplified DNA with short homology arms.
Yu, D. et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA 97, 5978–5983 (2000).The first use of a defective λ-prophage for recombineering.
Muyrers, J. P., Zhang, Y., Testa, G. & Stewart, A. F. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27, 1555–1557 (1999).The first use of ET cloning for BAC modification.
Muyrers, J. P., Zhang, Y. & Stewart, A. F. Techniques: recombinogenic engineering—new options for cloning and manipulating DNA. Trends Biochem. Sci. 26, 325–331 (2001).
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J. & Stahl, F.W. The double-strand-break repair model for recombination. Cell 33, 25–35 (1983).
Lafontaine, D. & Tollervey, D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24, 3469–3471 (1996).
Yamamoto, T., Moerschell, R. P., Wakem, L. P., Ferguson, D. & Sherman, F. Parameters affecting the frequencies of transformation and co-transformation with synthetic oligonucleotides in yeast. Yeast 8, 935–948 (1992).
Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F. & Cullin, C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329–3330 (1993).
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Kumar, A. & Snyder, M. Emerging technologies in yeast genomics. Nature Rev. Genet. 2, 302–312 (2001).
Storck, T., Kruth, U., Kolhekar, R., Sprengel, R. & Seeburg, P. H. Rapid construction in yeast of complex targeting vectors for gene manipulation in the mouse. Nucleic Acids Res. 24, 4594–4596 (1996).
Wilson, S. M. et al. The status of voltage-dependent calcium channels in α1E knock-out mice. J. Neurosci. 20, 8566–8571 (2000).
Wattler, S., Kelly, M. & Nehls, M. Construction of gene targeting vectors from λ KOS genomic libraries. Biotechniques 26, 1150–1156, 1158, 1160 (1999).
Bradshaw, M. S., Bollekens, J. A. & Ruddle, F. H. A new vector for recombination-based cloning of large DNA fragments from yeast artificial chromosomes. Nucleic Acids Res. 23, 4850–4856 (1995).
Bhargava, J. et al. Direct cloning of genomic DNA by recombinogenic targeting method using a yeast–bacterial shuttle vector, pClasper. Genomics 62, 285–288 (1999).
Pilarksi, L. M. & Egan, J. B. Role of DNA topology in transcription of coliphage λ in vivo. II. DNA topology protects the template from exonuclease attack. J. Mol. Biol. 76, 257–266 (1973).
Wackernagel, W. Genetic transformation in E. coli: the inhibitory role of the recBC DNase. Biochem. Biophys. Res. Commun. 51, 306–311 (1973).
Wackernagel, W. & Radding, C. M. Transfection by half molecules and inverted molecules of λ DNA: requirement for exo and -promoted recombination. Virology 52, 425–432 (1973).
Cosloy, S. D. & Oishi, M. The nature of the transformation process in Escherichia coli K12. Mol. Gen. Genet. 124, 1–10 (1973).
Benzinger, R., Enquist, L. W. & Skalka, A. Transfection of Escherichia coli spheroplasts. V. Activity of recBC nuclease in rec+ and rec− spheroplasts measured with different forms of bacteriophage DNA. J. Virol. 15, 861–871 (1975).
Oishi, M. & Cosloy, S. D. The genetic and biochemical basis of the transformability of Escherichia coli K12. Biochem. Biophys. Res. Commun. 49, 1568–1572 (1972).
Cosloy, S. D. & Oishi, M. Genetic transformation in Escherichia coli K12. Proc. Natl Acad. Sci. USA 70, 84–87 (1973).
Lloyd, R. G. & Buckman, C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164, 836–844 (1985).
Jasin, M. & Schimmel, P. Deletion of an essential gene in Escherichia coli by site-specific recombination with linear DNA fragments. J. Bacteriol. 159, 783–786 (1984).
Winans, S. C., Elledge, S. J., Krueger, J. H. & Walker, G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J. Bacteriol. 161, 1219–1221 (1985).
Bubeck, P., Winkler, M. & Bautsch, W. Rapid cloning by homologous recombination in vivo. Nucleic Acids Res. 21, 3601–3602 (1993).
Oliner, J. D., Kinzler, K. W. & Vogelstein, B. In vivo cloning of PCR products in E. coli. Nucleic Acids Res. 21, 5192–5197 (1993).
Smith, G. R. Homologous recombination in E. coli: multiple pathways for multiple reasons. Cell 58, 807–809 (1989).
Dabert, P. & Smith, G. R. Gene replacement with linear DNA fragments in wild-type Escherichia coli: enhancement by Chi sites. Genetics 145, 877–889 (1997).
Anderson, D. G. & Kowalczykowski, S. C. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11, 571–581 (1997).
Kowalczykowski, S. C., Dixon, D. A., Eggleston, A. K., Lauder, S. D. & Rehrauer, W. M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58, 401–465 (1994).
Myers, R. S., Stahl, M. M. & Stahl, F. W. Chi recombination activity in phage λ decays as a function of genetic distance. Genetics 141, 805–812 (1995).
Jessen, J. R. et al. Modification of bacterial artificial chromosomes through chi-stimulated homologous recombination and its application in zebrafish transgenesis. Proc. Natl Acad. Sci. USA 95, 5121–5126 (1998).This paper describes a BAC-modification system that can be used in wild-type E. coli cells.
Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P. & Kushner, S. R. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171, 4617–4622 (1989).
O'Connor, M., Peifer, M. & Bender, W. Construction of large DNA segments in Escherichia coli. Science 244, 1307–1312 (1989).
Yang, X. W., Model, P. & Heintz, N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nature Biotechnol. 15, 859–865 (1997).The first use of RecA-mediated recombination for BAC modification.
Imam, A. M. et al. Modification of human β-globin locus PAC clones by homologous recombination in Escherichia coli. Nucleic Acids Res. 28, E65 (2000).
Barbour, S. D., Nagaishi, H., Templin, A. & Clark, A. J. Biochemical and genetic studies of recombination proficiency in Escherichia coli. II. Rec+ revertants caused by indirect suppression of rec-mutations. Proc. Natl Acad. Sci. USA 67, 128–135 (1970).
Clark, A. J. et al. Genes of the RecE and RecF pathways of conjugational recombination in Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 49, 453–462 (1984).
Hall, S. D., Kane, M. F. & Kolodner, R. D. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J. Bacteriol. 175, 277–287 (1993).
Clark, A. J., Satin, L. & Chu, C. C. Transcription of the Escherichia coli recE gene from a promoter in Tn5 and IS50. J. Bacteriol. 176, 7024–7031 (1994).
Carter, D. M. & Radding, C. M. The role of exonuclease and β protein of phage λ in genetic recombination. II. Substrate specificity and the mode of action of λ exonuclease. J. Biol. Chem. 246, 2502–2512 (1971).
Little, J. W., Lehman, I. R. & Kaiser, A. D. An exonuclease induced by bacteriophage λ. I. Preparation of the crystalline enzyme. J. Biol. Chem. 242, 672–678 (1967).
Muniyappa, K., Shaner, S. L., Tsang, S. S. & Radding, C. M. Mechanism of the concerted action of recA protein and helix-destabilizing proteins in homologous recombination. Proc. Natl Acad. Sci. USA 81, 2757–2761 (1984).
Takahashi, N. & Kobayashi, I. Evidence for the double-strand break repair model of bacteriophage λ recombination. Proc. Natl Acad. Sci. USA 87, 2790–2794 (1990).
Murphy, K. C. λ Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol. 173, 5808–5821 (1991).
Murphy, K. C., Campellone, K. G. & Poteete, A. R. PCR-mediated gene replacement in Escherichia coli. Gene 246, 321–330 (2000).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Feiss, M., Siegele, D. A., Rudolph, C. F. & Frackman, S. Cosmid DNA packaging in vivo. Gene 17, 123–130 (1982).
Sergueev, K., Yu, D., Ausin, S. & Court, D. Cell toxicity caused by products of the pL operon of bacteriophage λ. Gene (in the press).
Lee, E. C. et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 (2001).This paper describes several modifications to the defective prophage system, and shows the use of the system for making cre -expressing BAC transgenic lines and for subcloning DNA fragments from BACs.
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 21, 70–71 (1999).
DiLeone, R. J., Russell, L. B. & Kingsley, D. M. An extensive 3′ regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics 148, 401–408 (1998).
Buchholz, F., Angrand, P. O. & Stewart, A. F. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nature Biotechnol. 16, 657–662 (1998).
Muyrers, J. P. et al. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 1, 239–243 (2000).This paper reports a simple and flexible two-step approach based on ET recombination, which allows point mutations, small insertions or deletions to be introduced into BACs without leaving any other residual change in the recombinant product.
Blomfield, I. C., Vaughn, V., Rest, R. F. & Eisenstein, B. I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5, 1447–1457 (1991).
Zhang, Y., Muyrers, J. P., Testa, G. & Stewart, A. F. DNA cloning by homologous recombination in Escherichia coli. Nature Biotechnol. 18, 1314–1317 (2000).In this study, ET recombination was used for the directed cloning and subcloning of DNA from complex mixtures and from target molecules that were resident in E. coli hosts.
Bhargava, J. et al. pPAC–ResQ: a yeast–bacterial shuttle vector for capturing inserts from P1 and PAC clones by recombinogenic targeted cloning. Genomics 56, 337–339 (1999).
Ellis, H. M., Yu, D., DiTizio, T. & Court, D. L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl Acad. Sci. USA 98, 6742–6746 (2001).This paper shows that phage-encoded Beta protein and short single-stranded DNA oligos can be used to mutagenize or repair the E. coli chromosome with high efficiency.
Swaminathan, S. et al. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis 29, 14–21 (2001).This study shows that mouse BACs can be modified at high efficiencies with short, single-stranded oligos and Beta protein. It also describes a simple PCR-based approach for detecting modified BACs.
Yamamoto, T., Moerschell, R. P., Wakem, L. P., Komar-Panicucci, S. & Sherman, F. Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics 131, 811–819 (1992).
Moerschell, R. P., Das, G. & Sherman, F. Transformation of yeast directly with synthetic oligonucleotides. Methods Enzymol. 194, 362–369 (1991).
Angrand, P. O., Daigle, N., Van der Hoeven, F., Scholer, H. R. & Stewart, A. F. Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res. 27, E16 (1999).
Hill, F. et al. BAC trimming: minimizing clone overlaps. Genomics 64, 111–113 (2000).
Ioannou, P. A. et al. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nature Genet. 6, 84–89 (1994).
Shizuya, H. et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA 89, 8794–8797 (1992).
Schedl, A. et al. A method for the generation of YAC transgenic mice by pronuclear microinjection. Nucleic Acids Res. 21, 4783–4787 (1993).
Gnirke, A., Huxley, C., Peterson, K. & Olson, M. V. Microinjection of intact 200- to 500-kb fragments of YAC DNA into mammalian cells. Genomics 15, 659–667 (1993).
Giraldo, P. & Montoliu, L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 10, 83–103 (2001).
Antoch, M. P. et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell 89, 655–667 (1997).
Peterson, K. R., Clegg, C. H., Li, Q. & Stamatoyannopoulos, G. Production of transgenic mice with yeast artificial chromosomes. Trends Genet. 13, 61–66 (1997).
Muyrers, J. P., Zhang, Y., Buchholz, F. & Stewart, A. F. RecE/RecT and Redα/Redβ initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 14, 1971–1982 (2000).
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- PHAGE-BASED RECOMBINATION
-
Bacteria, such as Escherichia coli, encode their own homologous recombination systems. Viruses or phages that inhabit bacteria also often carry their own recombination functions, which can work with, or independently of, the bacterial recombination functions.
- BACTERIAL ARTIFICIAL CHROMOSOME
-
(BAC). A cloning vector derived from a single-copy F-plasmid of Escherichia coli. It carries the F replication and partitioning systems that ensure low-copy number and faithful segregation of plasmid DNA to daughter cells. Large genomic fragments can be cloned into F-type plasmids, making them useful for constructing genomic libraries.
- PLASMID
-
An autonomously replicating DNA that is often marked with a gene that encodes drug resistance, which allows selection for cells that carry the plasmid.
- HIS3
-
A yeast selectable marker that encodes an enzyme required for histidine (His) biosynthesis. HIS3 yeast mutants cannot grow in media without His. On HIS-deficient medium, recombinants that restore the wild-type gene are able to grow again.
- GAP REPAIR
-
A linear plasmid vector (gapped vector) can be circularized by homologous recombination between its ends and a target DNA.
- SHUTTLE VECTOR
-
A plasmid that can be moved from one species to another, such as plasmids that contain origins of replication for both yeast and bacterial hosts.
- RECA
-
RecA is central to recombination in Escherichia coli, and all organisms have RecA homologues. It allows two homologous DNAs to find each other, and to trade DNA strands by binding to a single-stranded region in one of the DNAs and by using that strand to search for its double-stranded DNA (dsDNA) homologue. It then binds to a homologue, causing the single strand to pair with its complement in the dsDNA, displacing the identical strand of the duplex and generating a key intermediate in the recombination process.
- POSITIVE–NEGATIVE SELECTION
-
When the presence of a cassette is positively selected for, for example by drug resistance, and then negatively selected for, by eliminating cells that express a second selectable marker.
- DH10B
-
A strain of Escherichia coli that has been modified and selected to accept large BAC clones by transformation. DH10B is defective for RecA recombination.
- RAC PROPHAGE
-
Escherichia coli and other bacteria contain, in their chromosomes, remnants of viruses or prophages, such as Rac in E. coli, that often are defective and contain only a few genes of the original virus. Two Rac genes, recE and recT, encode homologous recombination functions, and are normally silent, but the sbcA mutation activates their constitutive expression.
- ARABINOSE
-
A simple five-carbon sugar metabolized by Escherichia coli, which is used as a chemical to induce and activate expression from the promoter pBAD.
- CRE
-
Cre is a site-specific recombinase that recognizes and binds to specific sites called loxP. Two loxP sites recombine at nearly 100% efficiency in the presence of Cre, allowing DNA cloned between two such sites to be removed by Cre-mediated recombination.
- FLPE
-
Flpe is a genetically enhanced, site-specific Flp recombinase that recognizes and binds to FRT sites.
- DY380
-
A derivative of DH10B. The defective λ-prophage, used to express the red and gam functions, has been moved into the chromosome of this strain.
- SACB
-
sacB encodes the SacB protein, which converts sucrose into a toxic form that kills bacteria. This can be used in negative selection for the sacB gene.
- FLAG TAG
-
A short peptide sequence that is added to a protein to allow the protein to be recognized by antibodies raised against the flag tag.
Rights and permissions
About this article
Cite this article
Copeland, N., Jenkins, N. & Court, D. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2, 769–779 (2001). https://doi.org/10.1038/35093556
Issue Date:
DOI: https://doi.org/10.1038/35093556
This article is cited by
-
Rapid Rescue of Goose Astrovirus Genome via Red/ET Assembly
Food and Environmental Virology (2024)
-
Generation of pulsatile ERK activity in mouse embryonic stem cells is regulated by Raf activity
Scientific Reports (2023)
-
dCas9-based gene editing for cleavage-free genomic knock-in of long sequences
Nature Cell Biology (2022)
-
A fast Myosin super enhancer dictates muscle fiber phenotype through competitive interactions with Myosin genes
Nature Communications (2022)
-
Resolving the structure of phage–bacteria interactions in the context of natural diversity
Nature Communications (2022)