Abstract
A Streptococcus dysgalactiae subsp. equisimilis (SDSE) strain WCHSDSE-1, which caused an outbreak of tonsillopharyngitis among healthcare workers in China, was subjected to genome sequencing and analysis. WCHSDSE-1 belongs to the Lancefield group G, emm type stG211.1 and sequence type 44. WCHSDSE-1 has virulence factors for adherence, impairing the recruitment of neutrophils to infection sites and toxins including streptolysins O and S and exotoxin G. WCHSDSE-1 has a 45.4-kb element resembling a conjugative transposon. This element is absent from other known SDSE genomes and contains the macrolide-resistant gene erm(B). Conjugative transfer of erm(B) was not successful in mating experiments, suggesting that the element might have lost its ability of conjugation. An almost identical element, which contains the tetracycline-resistant gene tet(M) instead of erm(B), is present on the genome of Filifactor alocis ATCC 35896. The boundaries and insertion sites of the two elements were identified and both were flanked by a 3-bp direct repeat, which is characteristic of transposition. In conclusion, the spectrum of virulence factors of WCHSDSE-1 is similar to other SDSE strains causing invasive diseases. WCHSDSE-1 possesses a new transposable element encoding macrolide resistance, which could pick up different resistance genes and could be transferred across species in oral microflora.
Similar content being viewed by others
Introduction
Streptococcus dysgalactiae subsp. equisimilis (SDSE) belongs to β-hemolytic streptococci. Although it has long been considered as a nonpathogenic colonizer of human throat and skin, SDSE is now recognized a causative agent of both human and livestock infections1. Like Streptococcus pyogenes, a well-known pathogen of group A streptococci, SDSE can cause a wide spectrum of human diseases from localized infection such as pharyngitis and pyoderma to invasive life-threatening infections such as bacteremia and toxin-mediated streptococcal toxic shock syndromes1. According to a widely-adopted classification scheme2, SDSE is classified into Lancefield group G (about 3/4), C (about 1/4) or A or L (occasionally). Besides SDSE, group G streptococci also include Streptococcus anginosus, Streptococcus canis and Streptococcus intestinalis1. Several outbreaks of pharyngitis or tonsillopharyngitis have been attributed to group G streptococci3,4,5,6,7, although the isolates have not been identified to the species level in these reports.
In August 2013, an outbreak of tonsillopharyngitis caused by SDSE among 30 healthcare workers (HCWs) occurred in a municipal hospital in China. Group G streptococci were recovered from the throat swabs of all of the 30 HCWs. All streptococci isolates were further identified as SDSE and were found to belong to the same strain (see below). One of the 30 isolates was randomly selected for whole genome sequencing and is reported here.
Results and Discussion
All SDSE isolates belonged to the same ST44-stG211.1 strain
All 30 isolates were identified as SDSE and belonged to the same emm type (stG211.1), the same sequence type (ST44) and the same pulsotype (unpublished data). The above findings suggest that the isolates represented a single strain. Among the 30 isolates, one, designated WCHSDSE-1, was selected for whole genome sequencing.
Genome sequence of SDSE isolate WCHSDSE-1
A total of 5,068,234 reads were obtained from the genome sequencing of WCHSDSE-1 comprising 506,823,400 bases with a 39.5% GC content. Reads were assembled into 70 contigs that were ≥500 bp in length (N50 = 63,425 bp) and contained 2,083,980 bp nucleotides. WCHSDSE-1 contained 2,076 protein coding sequences (CDS), 60 tRNA and 1 tmRNA.
ST44 corresponds to multiple emm types
In silico multi-locus sequence typing (MLST) confirmed that WCHSDSE-1 belonged to ST44, a sequence type which has not been found in many countries but has been reported in India and Australia before8,9. A single ST may correspond to multiple emm types, which represent serospecificities of S. pyogenes and possibly of group G and C streptococci such as SDSE. In a previous study, ST44 of SDSE was associated with five emm types (stC36, stG245, stG480, stG6 and stGL265)9, none of which is the stG211.1 type seen here. A SDSE isolate of stG211.1 has been previously recovered from a patient with pharyngitis in Vellore, India in 2007 but the ST of the isolate was not provided (the CDC emm database, ftp://ftp.cdc.gov/pub/infectious_diseases/biotech/tsemm/stG211.1.sds). In addition, a previous investigation on group G SDSE showed significant clonal diversity among SDSE in Chinese children10 but none of the isolates were stG211.1. Therefore, we believe that this is the first report of a ST44-stG211.1 SDSE isolate, which appears to be uncommon.
WCHSDSE-1 is close to a SDSE from Sweden by phylogenetic analysis.
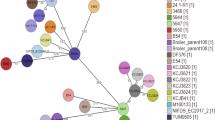
Based on the phylogenetic analysis, WCHSDSE-1 is most closely related to strain SK1250 (CCUG 45841; Fig.1), which belongs to Lancefield group A and emm type stG840 and was isolated from human throat in Sweden in 2001 ( http://www.straininfo.net/strains/330680/browser). The relative closeness between WCHSDSE-1 and SK1250 was also confirmed by in silico genome-to-genome comparisons (Table 1). Both phylogenetic and genome-to-genome analyses confirmed that the SDSE strains clustered together more closely in comparison to the S. dysgalactiae subsp. dysgalactiae (SDSD) strain ATCC 25957.
Phylogenetic tree of SDSE based on whole genome sequences.
Strains (GenBank accession number, genome statue [complete or draft]) are GGS_124 (AP010935, complete), ATCC 12394 (NC_017567, complete), RE378 (NC_018712, complete), AC-2713 (NC_019042, complete), 167 (NC_022532, complete), SK1250 (NZ_AFUL01000000, draft), ATCC 27957 (NZ_CM001076, only chromosome complete), SK1249 (NZ_AFIN01000000, draft), UT-5345 (NZ_LAKV01000000, draft), UT-5354 (NZ_LAKU01000000, draft), UT-SS957 (NZ_LAKT01000000, draft), UT-SS1069 (NZ_LAKS01000000, draft). ATCC 27957 is a SDSD strain and is included as an outgroup.
The virulence factor profile of WCHSDSE-1 is largely similar to other SDSE causing invasive diseases
The virulence factors of SDSE have been well characterized but were determined in strains causing invasive diseases rather than those causing tonsillopharyngitis. WCHSDSE-1 possessed a few virulence factors including: lmb, pavA, gfbA and gapC for adherence; scpA and scpB encoding complement proteases involved in an impaired recruitment of neutrophils to infection sites; determinants of streptolysins O and S and exotoxin G and genes encoding streptokinases (Table 2). Of note, superantigen G and streptolysin S genes that are regarded as the most important virulence factors causing invasive diseases11 are also present in WCHSDSE-1. On the other hand, the fnB gene, which encodes a fibronectin binding protein, is present in most SDSE strains causing invasive diseases but is absent from WCHSDSE-1. In contrast, WCHSDSE-1 possesses another fibronectin binding protein-encoding gene, gfbA, that is absent from most SDSE strains causing invasive diseases. The exact roles of fnB and gfbA of SDSE in causing invasive or non-invasive diseases remain undetermined but warrant further investigation. Most (79/88) of the proposed virulence factors, which have not been experimentally characterized, of the invasive strain GGS_124 are also present on WCHSDSE-1 (Table S1 in the Supplementary file). Among the 9 factors absent from WCHSDSE-1, two, i.e. sdn (encoding a deoxyribonuclease) and an unnamed gene (locus_tag 0157 of GGS_124, encoding a fimbrial subunit protein), are also absent from all other SDSE strains and therefore represent strain-specific factors for strain GGS_124. The remaining seven factors are either absent from at least one strain causing invasive diseases or present in some non-invasive strains. The spectrum of virulence factors of WCHSDSE-1 is therefore largely similar to SDSE strains causing invasive diseases (Table 2). Based on analysis here, invasive and non-invasive strains can not be distinguished by the spectrum of virulence factors.
As virulence factors may be carried by prophages, sequences belong to prophages were predicted from the genome sequence of WCHSDSE-1. Two prophages were identified, one of which is 37.6 kb and shows 94% identity (67% coverage) with the phage 3396 in a SDSE isolate (GenBank accession no. EF207558)12. Another prophage is 38.2 kb and shows 96% identity (45% coverage) with the phage T12 in Streptococcus pyogenes (GenBank accession no. KM289195). Of note, none of the virulence factors identified are present on either of the two prophages.
The macrolide-resistant gene erm(B) was carried by a new transposable element in WCHSDSE-1
Isolate WCHSDSE-1 haboured only one known antimicrobial resistance gene, which is erm(B). erm(B) is able to mediate resistance to macrolides, clindamycin and streptogramins B13. WCHSDSE-1 was susceptible to penicillin, cefotaxime, levofloxacin and vancomycin but was resistant to erythromycin, azithromycin, clindamycin and tetracycline. The presence of erm(B) corresponds to the macrolide and clindamycin resistance phenotype of WCHSDSE-1.
When the genetic context of erm(B) was examined, we found that a large 45.4-kb region containing erm(B) in WCHSDSE-1 was absent from other SDSE genomes. However, an almost identical region is present on the genome of Filifactor alocis ATCC 35896 (GenBank accession number CP002390). F. alocis is an oral Gram-positive anaerobic rod that can cause periodontal diseases14. However, the region of F. alocis ATCC 35896 has a 14.1-kb sequence containing the tetracycline resistance gene tet(M) in place of the 1.5-kb sequence containing erm(B) seen in isolate WCHSDSE-1 (Fig. 2). This 45.4-kb region of isolate WCHSDSE-1 appears to be a conjugative transposon as it contains genes encoding the site-specific recombinase, a relaxase-encoding gene (nicK), an origin of transfer (oriT; ACCCCCCGTATCTAACAGGGGGGT [inverted repeats are underlined], identical to that of the well-characterized conjugative transposon Tn916) upstream of nicK and a gene encoding conjugal transfer coupling protein. Of note, there are three recombinase-encoding genes, which cluster together (Fig. 2) and therefore it is reasonable to hypothesize that they may encode a single recombinase through frameshift. Mating experiments were performed to examine whether the putative transposon was conjugative. However, despite repeated attempts, conjugative transfer of erm(B) to the recipient strain was not detected. This suggests that the putative transposon carrying erm(B) in strain WCHSDSE-1 may have lost its ability of conjugation or the conjugation occurred in a rate lower than that could be detected in the mating experiments.
The conjugative transposon-like elements in WCHSDSE-1 and F. alocis ATCC 35896.
Different regions of the two elements are indicated by dotted lines. Boundaries of the transposon are indicated by black poles and the 3-bp DR (TGG) are shown. Genes and their product or function are listed: traE, conjugal coupling protein; traC, type-IV secretion system protein; iap, endopeptidase p60 precursor; topB, DNA topoisomerase 3; helicase gene, helicase/DNA methylase; rlx, relaxase/mobilisation nuclease domain protein; irtA, iron import ATP-binding/permease protein; msbA, putative ABC transporter ATP-binding protein; ecfT, energy-coupling factor transporter transmembrane protein; ykoD, putative HMP/thiamine import ATP-binding protein; ftsK, DNA translocase; nicK, relaxase; erm, macrolide resistance; rec, recombinase; tcpC, conjugative transposon protein; tet(M), tetracycline resistance; int, integrase; other genes are of unknown function. The scale in bp is shown.
This putative transposon was present in a single copy in both isolate WCHSDSE-1 and F. alocis ATCC 35896. Comparisons to other SDSE genome sequences allowed us to determine the exact boundary and the insertion site of this putative transposon. A 3-bp direct repeat (DR, Fig. 2) was identified flanking the transposon in isolate WCHSDSE-1 and the corresponding element in F. alocis ATCC 35896. In WCHSDSE-1, this putative transposon was inserted into a spacer region between two hypothetical open reading frames of unknown function (locus tags GGS_0498 and GGS_0499 in strain RE378; SDSE167_0576 and SDSE167_0577 in strain 167; SDEG_0522 and SDEG_0523 in strain GGS_124). The boundary of the putative transposon identified from isolate WCHSDSE-1 was also verified in F. alocis ATCC 35896. The putative transposon is also flanked by a 3-bp DR (Fig. 2) in F. alocis ATCC 35896. The evidence of transposition characterized by the presence of 3-bp DR suggests that the putative transposons in strain WCHSDSE-1 and F. alocis ATCC 3589 were truly transposable elements. The presence of highly similar transposable elements present in two species of human oral microflora suggests that interspecies transfer of a common transposon might have occurred among oral microflora.
Conclusions
We report the genome sequence of a stG211.1-ST44 SDSE strain WCHSDSE-1 causing tonsillopharyngitis. Isolate WCHSDSE-1 had a nearly identical spectrum of virulence factors with SDSE strains causing invasive diseases, suggesting that strains causing invasive and non-invasive diseases may not be distinguished by the presence or absence of certain virulence factors. A new putative transposon encoding macrolide resistance was identified. This putative transposon is likely to be acquired from species other than SDSE in the oral microflora and is able to pick up different antimicrobial resistance genes.
Methods
Species identification was performed by partially sequencing the 16S rRNA gene as described previously15. Three methods were used for strain typing: the emm (encoding M protein) genotyping scheme ( http://www.cdc.gov/streplab/protocol-emm-type.html), MLST ( http://sdse.mlst.net/) and pulsed field gel electrophoresis (PFGE)16.
Genomic DNA of isolate WCHSDSE-1 was prepared using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and was subjected to whole genome sequencing with a ca. 100 × coverage using the Hiseq 2500 Sequencer (Illumina, San Diego, CA, USA) following the manufacturer’s protocol at the Beijing Genomics Institute. Reads were assembled into contigs using the Spades program17 and the Prokka program18 was employed for annotating the genomic sequence. In silico MLST was performed using the MLST tool of Centre for Genomic Epidemiology. Prophages were predicted using the Prophinder program19, while known antimicrobial resistance genes were predicted using the ResFinder tool of Centre for Genomic Epidemiology.
A total of 11 SDSE genomes (5 complete and 6 draft versions) were available in GenBank ( http://www.ncbi.nlm.nih.gov/genome/genomes/823; accessed on June 17, 2015). The phylogenetic relatedness of WCHSDSE-1 and the 11 SDSE strains was investigated based on genome sequences by the Harvest suite20 and SDSD type strain ATCC 2795720 was included as an outgroup. In silico inter-genomic comparisons between WCHSDSE-1 and the 11 SDSE strains were performed using GGDC (formula 2) which mimics DNA-DNA hybridization (DDH)21. Potential unique genes were identified using the Gegenees program22 with a stringent threshold of a 0.9 or higher score generated by Gegenees.
A dataset of virulence factors of SDSE was established by retrieving the corresponding sequences (Table 2) from the Virulence Factors of Pathogenic Bacteria database (VFDB, http://www.mgc.ac.cn/cgi-bin/VFs/) and the review of Brandt et al.1 An additional dataset was compiled using the 88 known or putative virulence factors of SDSE strain GGS_124, which was demonstrated to cause streptococcal toxic shock syndrome23. The assembled contigs of WCHSDSE-1 were aligned against the datasets using the BLAST program. The presence of a virulence factor in WCHSDSE-1 was defined using the cutoff value ≥60% coverage and ≥70% nucleotide identity.
In vitro susceptibility of isolate WCHSDSE-1 to β-lactams (penicillin and cefotaxime), clindamycin, macrolides (erythromycin and azithromycin), levofloxacin, tetracycline and vancomycin were determined by disk diffusion following the recommendations of CLSI24.
Both filter-based25,26 and broth-based mating methods27 were used for conjugation experiments as described previously. A levofloxacin-resistant but erythromycin-susceptible Streptococcus agalactiae clinical isolate was used as the recipient. Transconjugants were selected on agar plates containing 1 μg/ml erythromycin and 8 μg/ml levofloxacin.
Additional Information
Accession Codes: Sequence reads and the Whole Genome Shotgun project of WCHSDSE-1 was depositedinto DDBJ/EMBLGenBank under accession SRR2049024 and LDYC00000000,respectively. The newputative conjugation transposon was deposited into DDBJ/EMBL/GenBank under accession KT005459.
How to cite this article: Wang, X. et al. Genome sequence and virulence factors of a group G Streptococcus dysgalactiae subsp. equisimilis strain with a new element carrying erm(B). Sci. Rep. 6, 20389; doi: 10.1038/srep20389 (2016).
References
Brandt, C. M. & Spellerberg, B. Human infections due to Streptococcus dysgalactiae subspecies equisimilis. Clin Infect Dis 49, 766–772 (2009).
Vieira, V. V. et al. Genetic relationships among the different phenotypes of Streptococcus dysgalactiae strains. Int J Syst Bacteriol 48 Pt 4, 1231–1243 (1998).
Gerber, M. A. et al. Community-wide outbreak of group G streptococcal pharyngitis. Pediatrics 87, 598–603 (1991).
Hill, H. R. et al. Epidemic of pharyngitis due to streptococci of Lancefield group G. Lancet 2, 371–374 (1969).
McCue, J. D. Group G streptococcal pharyngitis. Analysis of an outbreak at a college. JAMA 248, 1333–1336 (1982).
Cohen, D., Ferne, M., Rouach, T. & Bergner-Rabinowitz, S. Food-borne outbreak of group G streptococcal sore throat in an Israeli military base. Epidemiol Infect 99, 249–255 (1987).
Stryker, W. S., Fraser, D. W. & Facklam, R. R. Foodborne outbreak of group G streptococcal pharyngitis. Am J Epidemiol 116, 533–540 (1982).
McMillan, D. J. et al. Population genetics of Streptococcus dysgalactiae subspecies equisimilis reveals widely dispersed clones and extensive recombination. PLoS One 5, e11741 (2010).
McMillan, D. J. et al. Recombination drives genetic diversification of Streptococcus dysgalactiae subspecies equisimilis in a region of streptococcal endemicity. PLoS One 6, e21346 (2011).
Yin, J. et al. Molecular characterization of group G Streptococcus dysgalactiae subsp. equisimilis recovered from patients and healthy people in China. Diagn Microbiol Infect Dis 72, 41–46 (2012).
Abdelsalam, M., Chen, S. C. & Yoshida, T. Dissemination of streptococcal pyrogenic exotoxin G (spegg) with an IS-like element in fish isolates of Streptococcus dysgalactiae. FEMS Microbiol Lett 309, 105–113 (2010).
Davies, M. R. et al. Phage 3396 from a Streptococcus dysgalactiae subsp. equisimilis pathovar may have its origins in streptococcus pyogenes. J Bacteriol 189, 2646–2652 (2007).
Maravic, G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr Drug Targets Infect Disord 4, 193–202 (2004).
Aruni, A. W. et al. Filifactor alocis - a new emerging periodontal pathogen. Microbes Infect, In press. doi: 10.1016/j.micinf.2015.1003.1011. (2015).
Lane, D. J. 16S/23S rRNA sequencing. In: In Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons (1991).
Stanley, J. et al. Molecular subtyping of prevalent M serotypes of Streptococcus pyogenes causing invasive disease. J Clin Microbiol 33, 2850–2855 (1995).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477 (2012).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Lima-Mendez, G., Van Helden, J., Toussaint, A. & Leplae, R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 24, 863–865 (2008).
Treangen, T. J., Ondov, B. D., Koren, S. & Phillippy, A. M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15, 524 (2014).
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P. & Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14, 60 (2013).
Agren, J., Sundstrom, A., Hafstrom, T. & Segerman, B. Gegenees: fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS One 7, e39107 (2012).
Shimomura, Y. et al. Complete genome sequencing and analysis of a Lancefield group G Streptococcus dysgalactiae subsp. equisimilis strain causing streptococcal toxic shock syndrome (STSS). BMC Genomics 12, 17 (2011).
CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S23. Clinical and Laboratory Standards Institute (2013).
Bellanger, X. et al. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol 191, 2764–2775 (2009).
Haenni, M. et al. Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae and S. uberis. Appl Environ Microbiol 76, 7957–7965 (2010).
Smyth, D. J. et al. Conjugative transfer of ICESde3396 between three β-hemolytic streptococcal species. BMC Res Notes 7, 521 (2014).
Acknowledgements
The work was supported by a grant from the National Natural Science Foundation of China (project no. 81222025) and a grant from Sichuan Bureau of Science, China (project no. 2013JQ0042). We are grateful for Yi Xie, Mei Kang and Yu Feng for their technical assistance. We appreciate Dr. Björn Espedido, University of Western Sydney, Australia for his critical reading and thoughtful comments. We acknowledge the use of the Streptococcus dysgalactiae subspecies equisimilis MLST database which is located at Imperial College London and is funded by the Wellcome Trust.
Author information
Authors and Affiliations
Contributions
Concept and design: Z.Z.; Acquisition of data: X.W., X.X., Z.Z.; Analysis/Interpretation of data: X.W., Z.Z.; Draft of manuscript: Z.Z.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, X., Zhang, X. & Zong, Z. Genome sequence and virulence factors of a group G Streptococcus dysgalactiae subsp. equisimilis strain with a new element carrying erm(B). Sci Rep 6, 20389 (2016). https://doi.org/10.1038/srep20389
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep20389
This article is cited by
-
Inter-species gene flow drives ongoing evolution of Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis
Nature Communications (2024)
-
Emergence of a Streptococcus dysgalactiae subspecies equisimilis stG62647-lineage associated with severe clinical manifestations
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.