Abstract
Abiotic factors are major determinants of soil animal distributions and their dominant role is pronounced in extreme ecosystems, with biotic interactions seemingly playing a minor role. We modelled co-occurrence and distribution of the three nematode species that dominate the soil food web of the McMurdo Dry Valleys (Antarctica). Abiotic factors, other biotic groups, and autocorrelation all contributed to structuring nematode species distributions. However, after removing their effects, we found that the presence of the most abundant nematode species greatly, and negatively, affected the probability of detecting one of the other two species. We observed similar patterns in relative abundances for two out of three pairs of species. Harsh abiotic conditions alone are insufficient to explain contemporary nematode distributions whereas the role of negative biotic interactions has been largely underestimated in soil. The future challenge is to understand how the effects of global change on biotic interactions will alter species coexistence.
Similar content being viewed by others
Introduction
The processes that structure biological communities have been investigated intensely in the last decades1,2,3,4 over a broad range of scales5,6,7,8. In recent years, the major goal has been to understand the relative roles of the different processes that structure ecological communities with a special emphasis on the balance between stochastic (e.g., dispersal dynamics) and deterministic (e.g., competition for shared resources, environmental filtering) processes4,9,10,11,12,13,14,15.
Despite much progress, most studies have focused on the aboveground component of terrestrial ecosystems. The belowground component has received far less attention but there have been studies addressing soil community structure at very broad scales16,17,18 as well as studies focusing on particular groups (e.g., bacteria, fungi, arthropods) at relatively local scales6,19,20: soil communities are structured at multiple spatial scales with local communities often consisting of species that are dispersal limited over relatively broad spatial scales21,22. Dispersal limitation and small population size of local and partially isolated communities imply that stochastic processes can interact with deterministic processes (e.g., the selection exerted by environmental variables) and contribute to community structure at multiple scales6,22,23,24,25.
One driver of soil community structure that remains poorly understood is biotic interactions. In aboveground communities, ecologists have been generating strong evidence that biotic interactions such as partitioning of resources within trophic levels play a fundamental role in structuring local communities15,26,27. In belowground communities, within trophic-group interactions have been shown to play a key role only for certain groups such as fungi28, whereas soil animals have been postulated to be less controlled by direct biotic interactions22. Still, soil is a very heterogeneous habitat even at fine spatial scales (e.g., < 100 cm) and it is well established that this heterogeneity promotes soil animal diversity29. Biotic interaction could structure soil animal communities via niche partitioning along environmental gradients30 as suggested by earlier investigators31. That is, species can spatially segregate and coexist at relatively broader scales (i.e., colonise different patches in the same landscape) if competition for resources is incompatible with their coexistence at local scales12. Falsifying this hypothesis as applied to soil ecosystems is very challenging because of the physical and chemical complexity of the soil habitat and its high biotic diversity, which includes aboveground–belowground linkages. To test for this, ideally one would need to control for all the abiotic, biotic, and historical factors (e.g., dispersal dynamics, biogeographical legacies, etc.) that affect the co-occurrence of species32.
To address this challenge in the present study, we targeted the soil ecosystem of the McMurdo Dry Valleys (MDVs) of Antarctica. These valleys represent the largest ice-free area of the Antarctic continent, host the driest and coldest soils on Earth and are part of the US National Science Foundation’s Long Term Ecological Research (LTER) network. The valleys support a simple soil community dominated by microbes and nematodes, both of which play major roles in regulating C and N cycling33,34,35 and coexist with a very few species of microarthropods, rotifers, and tardigrades. Species richness is indeed possibly the lowest on Earth and the harsh environmental conditions make it possible to identify the most important environmental determinants of species distribution (e.g., water availability)35,36. Thus, thanks to an inherently simple ecosystem structure, the MDVs offer a unique opportunity to test the role of biotic interactions in the assembly of soil communities. Current evidence suggests that the spatial distribution of the MDVs biota is driven by abiotic factors37. We aimed to quantify the role of biotic interactions relative to the roles of all the other factors we measured. We focused on the three most common species of nematodes that dominate the species-poor soil animal community. The three most common MDV nematode species are Plectus murrayi, Eudorylaimus antarcticus, and Scottnema lindsayae38,39,40,41, hereafter referred to by only their genus. Species from a fourth genus, Geomonhystera antarcticola, as well as another species of Plectus, P. frigophilus, are present but rare in the region42. The three common and dominant nematode species are known to have different environmental preferences and feeding strategies43,44. Scottnema is a generalist that feeds on bacteria and fungi and is found in high numbers in the extreme, dry, saline soils of the Dry Valleys42,45,46. Like Scottnema, Plectus is also bactivorous47, whereas Eudorylaimus is an omnivore that feeds on algae, cyanobacteria, and likely a variety of other metazoans35,47,48. In terms of their habitat preferences, Plectus and Eudorylaimus prefer higher levels of soil moisture than Scottnema35,48.
Given these feeding preferences and the the very high dispersal capability of these species, we hypothesised the following hierarchy of processes15: first, following dispersal, a certain species colonises an environmentally suitable patch, although this same patch can also be colonised or already inhabited by another species. Second, there are two possibilities: either feeding preferences of the two species do not overlap for at least one important resource (Eudorylaimus–Plectus; Eudorylaimus–Scottnema) or feeding preferences overlap (Scottnema–Plectus) for one important resource. If feeding preferences overlap, the two species compete for resources and might or might not coexist locally, depending on resource distribution. The outcome of competition will depend on abiotic conditions and resource supply/consumption, and each species will win different patches under different abiotic conditions or the two species could occasionally coexist in some patches. In any case, this process is expected to make species co-occur less often than expected by chance, that is, to segregate32,49.
In this study, we tested the null hypothesis that the three species of nematodes co-occur randomly. More specifically, we searched for evidence for the alternative hypothesis that Scottnema and Plectus may segregate/repel each other or be negatively correlated both because they have different abiotic requirements and/or interact negatively. Because Eudorylaimus could be preying upon Scottnema or Plectus (or both), we also tested the hypothesis that Eudorylaimus could be negatively correlated with these species. We found that abiotic factors alone are insufficient to explain contemporary nematode distributions and show that the role of negative biotic interactions has been largely underestimated in soil.
Results
Species distribution
Scottnema, Eudorylaimus, and Plectus were found, respectively, in 289, 222, and 50 out of 314 sampling points in which at least one nematode was found. At least two species co-occurred in 217 out of 314 locations. All three species co-occurred in only 30 locations. Scottnema and Plectus co-occurred 32 times out of the possible 50 points where Plectus (the less frequent species of the pair) was found. Scottnema and Eudorylaimus co-occurred 204 times out of the 222 possible times, and Plectus and Eudorylaimus co-occurred 41 times out of the 50 possible.
The C-score, an index that quantifies checkerboard distributions, show that species co-occurred less often than expected by chance. The index value was, in fact, significantly larger than the central tendency of the null distribution (95% confidence limits: 663–752), corresponding to an effect size of 6.4. Bayes pair-wise analysis showed that two out of the three possible species pairs co-occurred non-randomly (P ≪ 0.001): Scottnema and Plectus segregated (effect size = 4.86), whereas Scottnema and Eudorylaimus aggregated (effect size = −4.32). Residuals from binomial generalized linear mixed models (GLMMs) (i.e., no modelling of spatial autocorrelation) displayed spatial structure when mapped (Fig. 1). This was particularly evident for Scottnema and Plectus. Autocorrelagrams based on Moran’s I confirmed there is spatial structure for neighbourhoods of about 4000–5000 m radius or less. GLMM for Scottnema revealed a significant (P < 0.05) negative effect of Plectus: the presence of Plectus significantly decreased the probability of occurrence of Scottnema while there is no effect of Eudorylaimus on Scottnema (Table 1). Scottnema was also positively correlated with the microbial biomass and richness gradient as well as the abundance of arthropods (Supplementary Figure 1). Scottnema was also negatively correlated to the soil moisture gradient, which correlated positively with electrical conductivity (i.e., the less water the higher the concentration of ions), pH, C, and N (Supplementary Figure 1). Also, Scottnema was positively correlated with the distance-to-the-coast gradient (Supplementary Figure 1). We observed a significant positive effect of the microbial biomass gradient and significant negative effect of Scottnema on Plectus (Table 2). Eudorylaimus (Supplementary Table 1) was not affected by the other two nematode species. Instead, the probability of occurrence of Eudorylaimus positively correlated with the richness of microbes and negatively correlated with the salinity and elevation gradient.
Map of the standardised residuals of Scottnema, Plectus, and Eudorylaimus. Residuals were obtained from a binomial GLM where predictors were the biotic and abiotic factors listed in Tables 1 and 2. Black and grey circles are negative and positive residuals, respectively. Negative correlation between Scottnema and Plectus is already evident in these maps and given these are residuals, this correlation does not depend on any measured environmental or biotic variables
Multivariate analysis of species abundances
A multivariate linear regression approach based on redundancy analysis (RDA) and variance partitioning showed that abiotic and biotic gradients plus spatial autocorrelation altogether account for 37% of total variance in the abundance of the three nematode species. Interestingly, spatial autocorrelation accounted for a relatively large and significant (P ≪ 0.05) fraction of variation (14 %) after controlling for the other factors, whereas biotic and abiotic factors accounted for 9% and 2% of variance, respectively. The remaining variance (12%) was shared among the three sources of variation. Residuals of the three species from the overall RDA model showed significant negative correlation (Fig. 2) for the pairs Scottnema-Plectus (Pearson’s r = −0.59, P < 0.001) and Scottnema-Eudorylaimus (Pearson’s r = −0.60, P < 0.001) no correlation was observed for Plectus-Eudorylaimus (Pearson’s r = −0.04, P = 0.49).
Scatter plots of the RDA residuals of Scottnema, Plectus, and Eudorylaimus. Residuals were obtained from an RDA model, the predictors of which were the same biotic and abiotic factors listed in Tables 1 and 2. RDA was applied to Hellinger transformed abundance data. Negative correlation is clear for the pairs Scottnema–Plectus (Pearson’s r = −0.59, P < 0.001) and the pairs Scottnema–Eudorylaimus (Pearson’s r = −0.60, P < 0.001)
Discussion
Negative biotic interactions such as competition for resources have been postulated to play a minor role in structuring soil animal communities22 despite suggestions by earlier investigators that partitioning of resources could explain the exceptional species richness observed in soil31. It is generally accepted that contemporary patterns of Antarctic biodiversity and ecosystem functioning are driven primarily by abiotic factors50. Thus, as pointed out by Convey et al.51, tests of hypotheses and model development for better understanding of patterns of species abundance and distributions must be dominated by measurements of abiotic environmental variables. The relative roles of biotic interactions has, however, remained unclear and the nzTABS project (see Methods) provided us with an optimal framework for developing models and testing hypotheses about these relative roles given its vast spatial scale and breadth and depth of biotic and abiotic variables measured. Our analysis shows, indeed, that a relatively large and statistically significant fraction of the variation observed in nematode distribution is spatially structured but independent of spatial variation in abiotic factors.
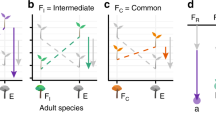
This variation could be explained by biotic interactions, stochastic processes (such as dispersal), or a combination of both. The existence of a large fraction of spatial variance independent of variation in abiotic factors is consistent with our conceptual framework (Fig. 3)12,15. In fact, in belowground communities part of this variation can be generated by the dispersal processes that link species regional pools to local communities19,21. These processes may well operate in terrestrial Antarctic ecosystems as inferred also by other authors25,52. Processes other than dispersal dynamics can, however, contribute to the fraction of spatial variation that is independent of abiotic factors and this might be particularly true for nematodes. Nematodes in the MDVs are not dispersal limited thanks to their life history and survival strategies, particularly cryoprotective dehydration46,53. Source-sink dynamics may thus support species in unfavourable patches, where conditions would usually cause species extinction after initial colonisation. High dispersal rates of nematodes can indeed support this dynamic, which is known as “mass effect” in metacommunity theory.
Conceptual model of the processes involved in the assembly of local belowground communities. The model reflects our modelling strategy. At large scales, dispersal determines whether a species colonises an environmentally suitable patch. Nematodes are not much dispersal limited. Environment is therefore filtering species from broader to smaller scales. In the patch, there might be local environmental heterogeneity but also interaction with other species, which either were already resident or may colonise the patch in a second moment. Species feeding preferences might not overlap (Eudorylaimus–Plectus; Eudorylaimus–Scottnema: see Gaussian curves of different colours), which makes local coexistence more probable. Or, species feeding preferences might overlap (Scottnema–Plectus), which might imply either local competitive exclusion and segregation or, in some cases local coexistence via stabilising mechanisms
Once a given species reaches a certain patch, abiotic and biotic conditions will determine whether or not the colonisation is successful (Fig. 1). This selection process is expected to create patterns of covariation in species distribution and abiotic/biotic conditions12,54. Two out of the three investigated species clearly showed this covariation and multivariate analysis showed that a significant fraction of variation in the overall assemblage was accounted for by abiotic factors. Scottnema is well adapted to dry, saline soil and known to be frequent and abundant at high elevations44,53. Our Scottnema GLMM confirms this. Eudorylaimus, too, is known to be well adapted to the Antarctic environment. However, studies of its ecology have highlighted that this species prefers moister conditions and relatively lower salinity than Scottnema, which might also be due to the fact that the Antarctic species of this genus can feed on algae35,44. Our Eudorylaimus GLMM is consistent with this notion. Also, the multivariate analysis demonstrates that part of the negative covariation between these two species can be due to their opposing responses to environmental gradients.
Abiotic factors thus act on two fundamental gradients in our system. The first is elevation, which together with distance to the coast affects temperature and water availability. The second is salinity, which is moderated by local topography but also partially depends on elevation and distance to the coast. Plectus was by far the least frequent and abundant of the three species, which is consistent with previous studies55. This species is typically associated with high moisture and its distribution is limited by the generally low soil moisture conditions of the MDVs42. Nevertheless, there are several patches where this species co-occurs with the other two species, as well as patches where Plectus was the only species present. Therefore, there are specific locations where Plectus can establish populations, at least in the short term, and perform even better than the other two species.
Two or more species often co-occurred in the same patch, and often at high densities. Eudorylaimus and Scottnema aggregated even though their relative abundances were negatively correlated. In general, the fact that the three species co-occur in a relatively high number of samples implies that these species could reach the same patch and cope with the abiotic and biotic conditions of this patch; Eudorylaimus might feed on a variety of food items (omnivorous), thus possibly avoiding competition with Plectus and Scottnema and potentially be indirectly favoured by C-cycling functions performed by Scottnema. Also, we cannot exclude the possibility that Eudorylaimus feed, at least occasionally, on Plectus and Scottnema, which could explain the negative correlation between the relative abundances of Scottnema and Eudorylaimus.
Our results suggest that Plectus and Scottnema could potentially compete for bacteria. That both these species feed on bacteria is a well established known factor, and our models show that bacterial richness and biomass positively correlate with frequency and abundance of both species. Plectus most likely feeds only on bacteria, whereas Scottnema is thought to feed on bacteria and yeast35,53. Under the hypothesis that these two species compete for one resource, our data empirically show that the two species have an ability to coexist locally under certain conditions. Thus, we can postulate the existence of stabilising niche differences that allow coexistence under specific conditions15. However, in most cases the two species do not co-occur and the analyses we have performed clearly suggest that the two species spatially segregate. This segregation cannot be fully explained by the numerous biotic and abiotic variables measured in the study. Given background knowledge and considering all the other factors taken into account in our statistical models, the observed pattern of segregation can be interpreted parsimoniously as a relative fitness difference15. When two species compete for a resource, relative fitness differences make one of the two species the best competitor for the resource (in this case bacteria) under a given set of abiotic and biotic conditions. However, if the conditions change, the other species can become the best competitor. Indeed, our modelling suggests that the two species respond to abiotic conditions differently: Scottnema is adapted to high salinity and low moisture and tolerate conditions under which Plectus would not survive (with or without competition). Plectus dominates only when soil moisture is much higher and salinity is lower than the average levels of MDVs. The two species have substantially different life strategies as Scottnema is stress tolerant and can survive in most MDV soil, being also able to grow optimally at low temperatures (optimum at ~10 C°)45, whereas Plectus flourishes at higher temperatures56.
In the past, biotic interactions have been considered a minor determinant of Antarctic species distribution in the belowground because there is little if any top down control (e.g., predators of microbial grazers are generally absent); and fundamental abiotic conditions such as availability of liquid water are extremely limiting37. More generally, the role of biotic interactions, especially competition, in determining distribution and coexistence of belowground invertebrates has been downplayed although not ruled out22,57. In this work, for the first time, we have shown that negative covariation between species of nematodes can be partly explained in terms of negative biotic interactions, besides the well-known effects of a range of abiotic factors, such as soil moisture and salinity. Future manipulative experiments will, however, have to directly test for these interactions explicitly.
The first implication of our findings is that biotic interactions could simply be underestimated in soil ecosystems because their effects are difficult to detect in the field and entangled with the effects of other, more macroscopic abiotic factors (e.g., large-scale gradients in salinity and moisture)27. Second, conditions for coexistence are more restrictive locally than globally30 as many species cannot always coexist locally but can colonise and win different patches in the same landscape, thereby still coexisting at broad scales if not at local scales. The interaction between the selection exerted by abiotic factors and biotic factors is expected to affect ecosystem functioning. In the case of the MDVs, Scottnema plays a crucial role in cycling soil organic C34. At the same time, climatic changes are happening at increasing rates in some areas of Antarctica58. These changes are drastically and negatively affecting the abundance and distribution of Scottnema34,58. We thus expect biotic interactions between Scottnema and other species to be altered by climatic changes because in many areas abiotic conditions are becoming less favourable to Scottnema. These changes have a great potential to affect ecosystem functioning35, in particular nutrient cycling, at larger scales34. More generally, our findings imply that the abiotic harshness of extreme environments alone is insufficient to explain the distribution of species across the landscape. Thus, biotic interactions may play a role in community assembly and subsequent ecosystem structure and functioning heretofore underestimated. The future challenge therefore is to quantify how environmental changes interfere with biotic interactions and affect the local and global coexistence of species and thus the effects of animal communities on ecosystem functions.
Methods
Study area and sampling design
This study was conducted in the context of the New Zealand Terrestrial Antarctic Biocomplexity Survey (nzTABS, http://nztabs.ictar.aq), which was initiated during the International Polar Year 2007–2008, and drew a diverse range of international expertise to profile the biology, geochemistry, geology, and climate of the Dry Valleys (Fig. 4). The study is among the most comprehensive landscape-scale biodiversity surveys undertaken and includes nearly all trophic components found in the Dry Valley ecosystem. A Geographic Information System (GIS) model including geological and geomorphological information and augmented by analyses of ALOS, LandSat, and MODIS satellite imagery, aerial photographs, and subsequent field mapping was used to divide the 220 km2 study area into 554 geographically and geologically distinct ice-free sectors (minimum area of 1.5 km2) referred to as landscape tiles. Tile boundaries were delineated where the combination of geographical and geological variables changed, which on average happened over a few km. On-the-ground environmental assessments were carried out in November 2008 to confirm the reliability of delineations. A total of 471 sites were chosen to encompass the entire range of geographical and geological heterogeneity in the sampling campaign. Sampling of soils and biological communities was carried out over two successive Austral summers (January 2009 and January 2010).
Map of the 220 km2 study area, in the McMurdo Dry Valleys. The area was divided into 554 geographically and geologically distinct ice-free sectors using remote-sensing data, and with geographical and geological variables validated on-the-ground assessments in November 2008. Eventually, 471 sites were sampled in January 2009 and January 2010 to encompass the entire range of geographical and geological heterogeneity in the area. Nematodes were found in 314 sites
Sampling abiotic and biotic variables
At each sample site, the top 10 cm of soil was collected using sterile techniques with a trowel from multiple spots within a 1 m2 area for the following subsamples: bulk soil (~400 g) with large pebbles ( > 2 cm diameter) removed aseptically and homogenised in a sterile 42 oz Whirl-Pak® bag; soil (~20 g) for moisture content measurement, subsampled from homogenised bulk soil into a sterile 15 mL centrifuge tube sealed with Parafilm®; soil (~300 g) for analysis of metazoans, stored in a sterile 18 oz Whirl-Pak® bag. The samples were staged in coolers in the field at ambient temperatures for up to 72 h prior to transport to the laboratory for further analysis.
Total soil moisture content was determined gravimetrically by the mass loss of soil heated to 105 °C for 48 h and recorded as percentage moisture content. For total organic carbon and total organic nitrogen contents, soil was air dried and ground in a ball mill to a fine homogenous powder. A 300 mg acidified aliquot of the homogenised soil was then analysed using a CE Elantech Flash EA 1112 Elemental Analyzer (Lakewood, NJ) at the Virginia Tech Ecosystem Analysis Laboratory59. Soil pH and conductivity were measured using the slurry method60. In brief, 10 mL of deionised water was added to a soil aliquot (2 mL) and mixed thoroughly. The pH and conductivity of the resulting slurry was measured using a Thermo Scientific Orion 4-Star Plus pH/Conductivity Meter (Thermo Scientific, Auckland, NZ).
A number of key environmental attributes were derived from satellite imagery and the custom digital elevation model created for the project, including basic topographic variables (such as elevation, slope, and aspect), surface soil temperature, a topographically derived “wetness index”, and distance to the coast. Soil surface temperatures were obtained from Landsat 7 ETM+ using band 6 at 60 m resolution, which captured the up-welling thermal infrared spectrum (in the 10.4–12.5 μm band). Replicated, summer Landsat 7-derived temperature data corresponding to locations of 45 on-the-ground temperature loggers (DS1921G iButtons, Maxim Integrated, San Jose, CA) were compared with records from the iButtons, and significant positive correlations between the two data sets were found61.
Nematodes, tardigrades, and rotifers were extracted from soils using the modified sugar centrifugation technique (see Supporting Information for details). Individual animals were identified and enumerated (both live and dead) using bright-field microscopy (Olympus CK40 Inverted Microscope, Olympus America Inc., Center Valley, PA). Nematodes were further identified into species, gender, and life stage (adult, juvenile) using Timm, Andrássy, and Boström et al.38,39,40,41. Population abundances were recorded as numbers of individuals per kg soil, corrected to oven-dry weight equivalent. The presence and abundance of flagellates, amoebae, and ciliates was also recorded. However, the data were not included in the analysis since reliable characterisations of protozoan abundance and diversity exceeded our logistical capability62.
Microarthropods (Acari and Collembola) were collected from the underside of small, flat, rocks within a 20 m radius of where the soil sample was taken. Individual animals were collected using an aspirator and preserved in 100% EtOH, enumerated, and identified using the approach described in63.
Total environmental DNA was extracted from soil samples using a CTAB protocol60 modified for the X-tractor Gene liquid handling robot (Corbett Life Sciences, Concorde, NSW, Australia). See Supplementary Methods for further details. The extracted environmental DNA was quality-checked and quantified as a surrogate for microbial biomass using Quant-iT Picogreen dsDNA reagent (Invitrogen, Auckland, New Zealand) on a FLUOstar optima fluorescence plate reader (BMG Laboratories, Offenburg, Germany. See Supplementary Methods for more details). Diversity of bacteria and fungi was estimated by automated ribosomal intergenic spacer analysis (ARISA64; see Supporting methods b in the Supporting Information document for details).
Soil respiration was measured by incubating 20 g (dry weight equivalent) samples of soils at 10 °C for 26–28 days in an miniaturised respiromotric chambers65 and the CO2 determined periodically by gas chemotography (Varion90 GC fitted with a thermal conductivity detector) as described by66.
Modelling
To collect evidence for our hypothesis that negative biotic interactions can structure the co-occurrence of the three species of nematodes, we first tested the general null hypothesis that species co-occur randomly (see Supporting Methods, Supplementary Figure 2). First, we used the species presence/absence matrix to calculate the C-score, an index that quantifies checkerboard distributions: species that do not co-occur very often produce a high index value and vice versa67. Second, we applied null model analysis49 to the C-score by using a randomisation scheme that preserved row and column totals (algorithm SIM9 in Gotelli49). This is the most effective randomisation approach to isolate non-random patterns caused by biological interactions because it randomises only species composition. The C-score plus the randomisation scheme we employed has been shown to lower the risk of false positives while maintaining good statistical power49. The null distribution of the C-score was obtained from 5000 random matrices. The central tendency of the null distribution was then compared with the observed C-score. The C-score was also calculated on a species-pair basis and tested following the method of Gotelli and Ulrich68 and the Fortran program Pairs69: this method builds confidence limits using the empirical Bayes approach. In null models, effect size was calculated as \(\frac{{ {\rm obs}. {\rm index} - {\rm exp}. {\rm index}}}{{ {\rm null}\, {\rm S.D.}}}\), where obs.index is the observed C-score, exp.index is the central tendency in the C-score null distribution, and null S.D. is the Standard Deviation of the C-score null distribution.
To isolate the relative effects of abiotic and biotic variables, we modelled the effect of the presence of one species on the other while statistically controlling for all other measured variables. We thus analysed the presence/absence distribution of each species using GLMM (spatial generalised linear mixed model70); with binomial error structure71,72. These models estimate the probability that a particular species colonises a certain patch and how measured abiotic and biotic variables together with the presence of the other two species of nematodes affected this probability. In order to avoid collinearity between predictors and model overfitting (i.e., too many and too correlated predictors), we applied principal component analysis (PCA) to the correlation matrix of abiotic and biotic variables to estimate major gradients in the system. We selected the first PCA axes that accounted for at least 2/3 of total variance73 and used these axes to quantify major abiotic and biotic gradients (see Supplementary Methods and Supplementary Figure 1 for details on how biotic and abiotic predictors were introduced in the models). GLMM models also allowed us to quantify the size of the effect of each variable on the occurrence of each species while controlling for spatial autocorrelation in species distribution. We used a spherical autocorrelation function74,75 to model spatial autocorrelation.
Recent studies have shown that co-occurrence data might reflect biotic interactions only partially76,77 and we thus completed the analysis with a multivariate modelling of the relative abundances of species. Specifically, we applied Hellinger transformation to the matrix of abundance of the three nematode species. This allowed us to correctly apply Redundancy Analysis, a multivariate form of linear regression73, to species abundance data78. The predictors used in RDA were the same as the predictors used in GLMMs. In addition, we used principal coordinate analysis of neighbour matrices (PCNM79); to account for spatial autocorrelation. Following80, we used a multivariate extension of the Akaike Information Criterion (AIC) to select the most parsimonious set of vectors accounting for the largest amount of autocorrelation in species distribution. Variance partitioning was calculated to quantify the amount of variation accounted for by each set of predictors (abiotic, biotic variables and PCNM eigenvectors, below called “spatial vectors”). Finally, RDA residuals of each of the three nematode species were correlated to the residuals of the other two species. GLMMs and RDA were run in R version 2.15.181 using the package MASS70 and vegan82.
Code availability
R codes used for Multivariate modelling and GLMM models are available as Supplementary Software 1 and 2.
Data availability
The full dataset is uploaded online as Supplementary Data 1.
References
Fargione, J., Brown, C. S. & Tilman, D. Community assembly and invasion: an experimental test of neutral versus niche processes. Proc. Natl Acad. Sci. USA 100, 8916–8920 (2003).
Chang, L.-W., Zelený, D., Li, C.-F., Chiu, S.-T. & Hsieh, C.-F. Better environmental data may reverse conclusions about niche‐and dispersal‐based processes in community assembly. Ecology 94, 2145–2151 (2013).
Nemergut, D. R. et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356 (2013).
Vellend, M. The Theory of Ecological Communities. (Princeton University Press, Princeton, NJ, USA, 2016).
Dornelas, M. Disturbance and change in biodiversity. Proc. R. Soc. B 365, 3719–3727 (2010).
Caruso, T. et al. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 5, 1406–1413 (2011).
Hortal, J., De Marco, P. Jr, Santos, A. & Diniz‐Filho, J. A. F. Integrating biogeographical processes and local community assembly. J. Biogeogr. 39, 627–628 (2012).
Mayfield, M. M. & Stouffer, D. B. Higher-order interactions capture unexplained complexity in diverse communities. Nat. Ecol. Evol. 1, 0062 (2017).
Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366 (2000).
Hubbell, S. P. The Unified Neutral Theory of Biodiversity and Biogeography. (Princeton University Press, Princeton, NJ, USA, 2001).
Hille Ris Lambers, J., Clark, J. S. & Beckage, B. Density-dependent mortality and the latitudinal gradient in species diversity. Nature 417, 732–735 (2002).
Leibold, M. A. et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol. Lett. 7, 601–613 (2004).
Dornelas, M., Connolly, S. R. & Hughes, T. P. Coral reef diversity refutes the neutral theory of biodiversity. Nature 440, 80–82 (2006).
Adler, P. B., HilleRisLambers, J. & Levine, J. M. A niche for neutrality. Ecol. Lett. 10, 95–104 (2007).
HilleRisLambers, J., Adler, P. B., Harpole, W. S., Levine, J. M. & Mayfield, M. M. Rethinking community assembly through the lens of coexistence theory. Annu. Rev. Ecol. Evol. Syst. 43, 227–248 (2012).
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A. & Cleveland, C. C. Global patterns in belowground communities. Ecol. Lett. 12, 1238–1249 (2009).
De Vries, F. T. et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 15, 1230–1239 (2012).
Wu, T., Ayres, E., Bardgett, R. D., Wall, D. H. & Garey, J. R. Molecular study of worldwide distribution and diversity of soil animals. Proc. Natl Acad. Sci. USA 108, 17720–17725 (2011).
Lindo, Z. & Winchester, N. Spatial and environmental factors contributing to patterns in arboreal and terrestrial oribatid mite diversity across spatial scales. Oecologia 160, 817–825 (2009).
Ramirez, K. S. et al. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proc. R. Soc. London B Biol. Sci. https://doi.org/10.1098/rspb.2014.1988 (2014).
Ettema, C. H. & Wardle, D. A. Spatial soil ecology. Trends Ecol. Evol. 17, 177–183 (2002).
Wardle, D. A. The influence of biotic interactions on soil biodiversity. Ecol. Lett. 9, 870–886 (2006).
Dumbrell, A. J., Nelson, M., Helgason, T., Dytham, C. & Fitter, A. H. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 4, 337–345 (2010).
Dumbrell, A. J., Nelson, M., Helgason, T., Dytham, C. & Fitter, A. H. Idiosyncrasy and overdominance in the structure of natural communities of arbuscular mycorrhizal fungi: is there a role for stochastic processes? J. Ecol. 98, 419–428 (2010).
Sokol, E. R., Herbold, C. W., Lee, C. K., Cary, S. C. & Barrett, J. Local and regional influences over soil microbial metacommunities in the Transantarctic Mountains. Ecosphere 4, 1–24 (2013).
Harpole, W. S. & Tilman, D. Non-neutral patterns of species abundance in grassland communities. Ecol. Lett. 9, 15–23 (2006).
Levine, J. M. & HilleRisLambers, J. The importance of niches for the maintenance of species diversity. Nature 461, 254–257 (2009).
Wardle, D. A. Communities and Ecosystems: Linking the Aboveground and Belowground Components. (Princeton University Press, Princeton, NJ, USA, 2002).
Nielsen, U. N. et al. The enigma of soil animal species diversity revisited: the role of small-scale heterogeneity. PLoS ONE 5, e11567 (2010).
Chase, J. M. & Leibold, M. A. Ecological Niches: Linking Classical and Contemporary Approaches. (Chicago University Press, Chicago, IL, USA, 2003).
Anderson, J. The enigma of soil animal species diversity. in Progress in Soil Zoology 51–58 (Springer, Prague, 1975).
Gotelli, N. J. & Ulrich, W. Statistical challenges in null model analysis. Oikos 121, 171–180 (2012).
Barrett, J. E. et al. Terrestrial ecosystem processes of Victoria Land, Antarctica. Soil Biol. Biochem. 38, 3019–3034 (2006).
Barrett, J. E., Virginia, R. A., Wall, D. H. & Adams, B. J. Decline in a dominant invertebrate species contributes to altered carbon cycling in a low-diversity soil ecosystem. Glob. Change Biol. 14, 1734–1744 (2008).
Wall, D. H. Global change tipping points: above- and below-ground biotic interactions in a low diversity ecosystem. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 2291–2306 (2007).
Bergstrom, D. M., Convey, P. & Huiskes, A. H. L. Trends in Antarctic Terrestrial and Limnetic Ecosystems: Antarctica as a Global Indicator. (Springer-Verlag, Dordrecht, 2006).
Hogg, I. D. et al. Biotic interactions in Antarctic terrestrial ecosystems: are they a factor? Soil Biol. Biochem. 38, 3035–3040 (2006).
Timm, R. Antarctic soil and freshwater nematodes from the McMurdo Sound region. Proc. Helminthol. Soc. Wash. 38, 42–52 (1971).
Andrássy, I. Nematodes in the sixth continent. J. Nematode Morphol. Syst. 1, 107–186 (1998).
Andrássy, I. Eudorylaimus species (Nematoda: Dorylaimida) of continental Antarctica. J. Nematode Morphol. Syst. 11, 11–11 (2008).
Boström, S., Holovachov, O. & Nadler, S. A. Description of Scottnema lindsayae Timm, 1971 (Rhabditida: Cephalobidae) from Taylor Valley, Antarctica and its phylogenetic relationship. Polar. Biol. 34, 1–12 (2011).
Adams, B. J., Wall, D. H., Virginia, R. A., Broos, E. & Knox, M. A. Ecological biogeography of the terrestrial nematodes of Victoria Land, Antarctica. Zookeys https://doi.org/10.3897/zookeys.419.7180 (2014).
Courtright, E. M., Wall, D. H. & Virginia, R. A. Determining habitat suitability for soil invertebrates in an extreme environment: the McMurdo Dry Valleys, Antarctica. Antarct. Sci. 13, 9–17 (2001).
Nielsen, U. N., Wall, D. H., Adams, B. J. & Virginia, R. A. Antarctic nematode communities: observed and predicted responses to climate change. Polar Biol. https://doi.org/10.1007/s00300-011-1021-2 (2011).
Overhoff, A., Freckman, D. W. & Virginia, R. A. Life cycle of the microbivorous Antarctic Dry Valley nematode Scottnema lindsayae (Timm 1971). Polar. Biol. 13, 151–156 (1993).
Nkem, J., Virginia, R., Barrett, J., Wall, D. & Li, G. Salt tolerance and survival thresholds for two species of Antarctic soil nematodes. Polar. Biol. 29, 643–651 (2006).
Yeates, G. W., Bongers, Td, De Goede, R., Freckman, D. & Georgieva, S. Feeding habits in soil nematode families and genera—an outline for soil ecologists. J. Nematol. 25, 315 (1993).
Shaw, E. A. et al. Stable C and N isotope ratios reveal soil food web structure and identify the nematode Eudorylaimus antarcticus as an omnivore–predator in Taylor Valley, Antarctica. Polar Biol. https://doi.org/10.1007/s00300-017-2243-8 (2018).
Gotelli, N. J. Null model analysis of species co-occurence patterns. Ecology 81, 2606–2621 (2000).
Convey, P. The influence of environmental characteristics on life history attributes of Antarctic terrestrial biota. Biol. Rev. 71, 191–225 (1996).
Convey, P. et al. The spatial structure of Antarctic biodiversity. Ecol. Monogr. 84, 203–244 (2014).
Chown, S. L. & Convey, P. Spatial and temporal variability across life’s hierarchies in the terrestrial Antarctic. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 2307–2331 (2007).
Adams, B. J. et al. The southernmost worm, Scottnema lindsayae (Nematoda): diversity, dispersal and ecological stability. Polar. Biol. 30, 809–815 (2007).
Cottenie, K. Integrating environmental and spatial processes in ecological community dynamics. Ecol. Lett. 8, 1175–1182 (2005).
Treonis, A. M., Wall, D. H. & Virginia, R. A. Invertebrate biodiversity in Antarctic Dry Valley soils and sediments. Ecosystems 2, 482–492 (1999).
Yeates, G. W., Scott, M. B., Chown, S. L. & Sinclair, B. J. Changes in soil nematode populations indicate an annual life cycle at Cape Hallett, Antarctica. Pedobiologia 52, 375–386 (2009).
Caruso, T., Trokhymets, V., Bargagli, R. & Convey, P. Biotic interactions as a structuring force in soil communities: evidence from the micro-arthropods of an Antarctic moss model system. Oecologia https://doi.org/10.1007/s00442-012-2503-9 (2012).
Nielsen, U. N. & Wall, D. H. The future of soil invertebrate communities in polar regions: different climate change responses in the Arctic and Antarctic? Ecol. Lett. 16, 409–419 (2013).
Barrett, J., Gooseff, M. & Takacs-Vesbach, C. Spatial variation in soil active-layer geochemistry across hydrologic margins in polar desert ecosystems. Hydrol. Earth Syst. Sci. 13, 2349-2358 (2009).
Lee, C. K., Barbier, B. A., Bottos, E. M., McDonald, I. R. & Cary, S. C. The inter-valley soil comparative survey: the ecology of Dry Valley edaphic microbial communities. ISME J. 6, 1046–1057 (2012).
Brabyn, L. et al. Accuracy assessment of land surface temperature retrievals from Landsat 7 ETM+in the Dry Valleys of Antarctica using iButton temperature loggers and weather station data. Environ. Monit. Assess. 186, 2619–2628 (2014).
Bamforth, S. S., Wall, D. H. & Virginia, R. A. Distribution and diversity of soil protozoa in the McMurdo Dry Valleys of Antarctica. Polar. Biol. 28, 756–762 (2005).
Stevens, M. I. & Hogg, I. D. Long‐term isolation and recent range expansion from glacial refugia revealed for the endemic springtail Gomphiocephalus hodgsoni from Victoria Land, Antarctica. Mol. Ecol. 12, 2357–2369 (2003).
Gardes, M. & Bruns, T. D. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118 (1993).
Heilmann, B. & Beese, F. Miniaturized method to measure carbon dioxide production and biomass of soil microorganisms. Soil Sci. Soc. Am. J. 56, 596–598 (1992).
Hopkins, D. & Ferguson, K. Substrate induced respiration in soil amended with different amino acid isomers. Appl. Soil Ecol. 1, 75–81 (1994).
Stone, L. & Roberts, A. The checkerboard score and species distributions. Oecologia 85, 74–79 (1990).
Gotelli, N. & Ulrich, W. The empirical Bayes approach as a tool to identify non-random species associations. Oecologia 162, 463–477 (2010).
Ulrich, W. Pairs - a FORTRAN program for studying pair-ise species associations in ecological matrices. http://www.home.umk.pl/~ulrichw/?Research:Software:Pairs (2008).
Venables, V. N. & Ripley, B. D. Modern Applied Statistics with S. (Springer-Verlag, New York, 2002).
McCullagh, P. & Nelder, J. Generalized Linear Models. (Chapman and Hall, London, 1989).
Quinn, G. P. & Keough, M. J. Experimental Design and Data analysis for Biologists. (Cambridge University Press, Cambridge, UK, 2002).
Legendre, P. & Legendre, L. Numerical Ecology. (Elsevier, Amsterdam, 1998).
Dormann, F. C. et al. Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30, 609–628 (2007).
Zuur, A. F., Ieno, E. N., Walker, N. J., Saveliev, A. A. & Smith, G. M. Mixed Effects Models and Extensions in Ecology with R. (Springer-Verlag, New York, 2009).
Barner Allison, K., Coblentz Kyle, E., Hacker Sally, D. & Menge Bruce, A. Fundamental contradictions among observational and experimental estimates of non‐trophic species interactions. Ecology 99, 557–566 (2018).
Freilich Mara, A., Wieters, E., Broitman Bernardo, R., Marquet Pablo, A. & Navarrete Sergio, A. Species co‐occurrence networks: Can they reveal trophic and non‐trophic interactions in ecological communities? Ecology 99, 690–699 (2018).
Legendre, P. & Gallagher, E. D. Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280 (2001).
Borcard, D., Legendre, P., Avois-Jacquet, C. & Tuomisto, H. Dissecting the spatial structure of ecological data at multiple scales. Ecology 85, 1826–1832 (2004).
Dray, S., Legendre, P. & Peres-Neto, P. R. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol. Modell. 196, 483–493 (2006).
R Development Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2009).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 1.15–4. http://CRAN.R-project.org/package=vegan (2009).
Acknowledgements
This research was supported by grants from the New Zealand Foundation for Research, Science and Technology (UOWX0710) to S.C.C. and T.G.A.G.; the New Zealand Marsden Fund to C.K.L. (UOW1003); the New Zealand Ministry of Business, Innovation and Employment to S.C.C., C.K.L., I.D.H. and B.C.S. (UOWX1401); and the United States National Science Foundation (ANT-0944556, ANT-0944560 to S.C.C. and LTER OPP 115245 to B.J.A., J.E.B. and D.H.W.); FP7 Marie Curie Actions (SENSE: Structure and Ecological Niche in the Soil Environment; EC FP7 - 631399 - SENSE) to T.C. We acknowledge contributions from all participants of nzTABS field expeditions, including J.C. Banks, L. Brabyn, C.M. Beard, M.J. Bentley, N. Carson, C. Colesie, Y. Cook, D.A. Cowan, N.J. Demetras, R. Gademann, I. Jones, K. Joy, D.C. Laughlin, I.R. McDonald, J.R. Quilez, A. de los Rios, P. Ross, U. Ruprecht, L. Sancho, R. Seppelt, T. Smith, A.D. Sparrow, M.I. Stevens, G.A. Stichbury, G. Tiao, and many others (in alphabetical order). We thank Antarctica New Zealand and the United States Antarctic Program for logistics support. Bishwo Adhikari, Karen Seaver, Zach Sylvain, and Breana Simmons provided field and lab assistance.
Author information
Authors and Affiliations
Contributions
T.C. conducted the modelling and statistical analysis. B.J.A., I.D.H., U.N., C.K.L., E.M.B., J.E.B. and D.W.H. performed sample processing and analysis. B.J.A., D.H.W. and U.N. performed taxonomic identification; S.C.C. and T.G.A.G. conceived the field study; C.K.L., S.C.C., T.G.A.G., I.D.H. and B.C.S. designed the field study and prepared funding proposals; C.K.L., S.C.C. and E.M.B. coordinated field expeditions. T.C. led the writing effort with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Caruso, T., Hogg, I.D., Nielsen, U.N. et al. Nematodes in a polar desert reveal the relative role of biotic interactions in the coexistence of soil animals. Commun Biol 2, 63 (2019). https://doi.org/10.1038/s42003-018-0260-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-018-0260-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.