Abstract
The Annonaceae, the largest family in the early-divergent order Magnoliales, comprises 107 genera and c. 2,400 species. Previous molecular phylogenetic studies targeting different taxa have generated large quantities of partially overlapping DNA sequence data for many species, although a large-scale phylogeny based on the maximum number of representatives has never been reconstructed. We use a supermatrix of eight chloroplast markers (rbcL, matK, ndhF, psbA-trnH, trnL-F, atpB-rbcL, trnS-G and ycf1) to reconstruct the most comprehensive tree to date, including 705 species (29%) from 105 genera (98%). This provides novel insights into the relationships of five enigmatic genera (Bocagea, Boutiquea, Cardiopetalum, Duckeanthus and Phoenicanthus). Fifteen main clades are retrieved in subfamilies Annonoideae and Malmeoideae collectively, 14 of which correspond with currently recognised tribes. Phoenicanthus cannot be accommodated in any existing tribe, however: it is retrieved as sister to a clade comprising the tribes Dendrokingstonieae, Monocarpieae and Miliuseae, and we therefore validate a new tribe, Phoenicantheae. Our results provide strong support for many previously recognised groups, but also indicate non-monophyly of several genera (Desmopsis, Friesodielsia, Klarobelia, Oxandra, Piptostigma and Stenanona). The relationships of these non-monophyletic genera—and two other genera (Froesiodendron and Melodorum) not yet sampled—are discussed, with recommendations for future research.
Similar content being viewed by others
Introduction
The Annonaceae are a pantropical family of flowering trees, shrubs and lianas, predominantly growing in tropical and subtropical lowland forests. It is the most species-rich family amongst the early-divergent magnoliids1, with 107 genera and c. 2,400 species currently recognised (Table 1). Previous molecular phylogenetic analyses2,3,4,5 have clarified backbone relationships within the family and hence contributed towards a framework for addressing higher-level infrafamilial taxonomy. A recently published phylogenetic study based on eight plastid markers and representatives of 94 genera2
was used to formally classify the Annonaceae into four subfamilies, Anaxagoreoideae, Ambavioideae, Annonoideae and Malmeoideae, and further subdivide these subfamilies into 14 tribes.
Since the publication of the family-wide phylogeny of Annonaceae2, several other molecular (and combined molecular-morphological) studies have made important contributions to our understanding of phylogenetic relationships and generic circumscriptions in various lineages, including: Disepalum 6, Friesodielsia-Monanthotaxis 7, Goniothalamus 8, Huberantha (as ‘Hubera’)9, Meiogyne 10, tribe Piptostigmateae11, Polyalthia-Monoon 12, Wangia 13 and Winitia 14. Despite these significant advances over the past decade, several genera (Bocagea, Boutiquea, Cardiopetalum, Duckeanthus, Froesiodendron, Melodorum and Phoenicanthus) remain unsampled due to difficulties in obtaining DNA of sufficient quality for phylogenetic reconstruction, and hence their systematic placements within the family remain unknown.
A robust phylogenetic framework based on a maximum number of Annonaceae representatives is invaluable for understanding the diversity, classification and evolution of the family. Numerous recently published phylogenetic studies have focused on specific genera and hence have incorporated differing taxon sampling; these studies have generated large quantities of partially overlapping sequence data, providing an opportunity for a wider family-level analysis addressing some gaps in our current knowledge of phylogenetic relationships.
In this study, we reconstruct the phylogeny of the Annonaceae based on a supermatrix of eight chloroplast loci and 749 accessions representing 705 species (29% of c. 2,400 currently recognised species) of 105 genera (98% of 107 currently accepted genera). The data matrix includes nearly four times as many species and representatives of 15 additional genera in comparison to the largest previous study (193 spp.)2. The aims of this study are: (i) to reconstruct the most comprehensive evolutionary tree of life for the Annonaceae available to date, providing a robust platform for future evolutionary studies; (ii) to determine the phylogenetic position of five genera (Bocagea, Boutiquea, Cardiopetalum, Duckeanthus and Phoenicanthus), which were not included in any previous molecular phylogenetic reconstructions; (iii) to assess the monophyletic status and phylogenetic relationships within each major clade, highlighting possible non-monophyly of genera and evaluating alternative resolutions to nomenclatural problems; (iv) to identify and discuss additional taxonomic problems that await resolution, including the phylogenetic placement and taxonomy of two genera, Froesiodendron and Melodorum, which have not been sampled yet; and (v) to provide an updated overview of currently recognised genera in the family (Table 1) with their species richness.
Results
The concatenated alignment for the dataset with 754 terminals consisted of 10,782 positions. The characteristics and best-fitting nucleotide substitution model for each data matrix are presented in Table 2. A summary of the best-scoring maximum likelihood (ML) tree showing the phylogenetic backbone of the Annonaceae is presented in Figs 1 and 2, with tips representing genera (or subdivisions of genera when not monophyletic). The entire tree with all 754 terminals is presented as Supplementary Figs S1–S9. An updated list of currently recognised genera in the Annonaceae is given in Table 1, with 107 genera and c. 2,400 species.
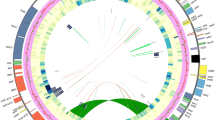
Skeletal representation of the best-scoring maximum likelihood tree inferred from a 754-accession dataset of eight chloroplast markers, showing topology of Anaxagoreoideae, Ambavioideae and Annonoideae. Tips represent genera or subdivisions when genera are not monophyletic. Higher taxon names appear to the right, marked with different background colours. ML bootstrap (BS) values ≥50, MP jackknife (JK) values ≥50 and Bayesian posterior probabilities (PP) values ≥0.5 are indicated at each node: BS /JK/ PP. -, represents clade support values <50%. Numbers in brackets show numbers of sampled taxon and supplementary figures of each lineage. Subfamily and tribe abbreviations: AN: Anaxagoreoideae; GU: Guatterieae; XY: Xylopieae. Flower morphology of the selected genera: (A) Anaxagorea luzonensis; (B) Lettowianthus stellatus; (C) Mkilua fragrans; (D) Duguetia confinis; (E) Fusaea longifolia; (F) Artabotrys hongkongensis; (G) Goniothalamus repevensis; (H) Anonidium floribundum; (I) Monodora myristica; (J) Isolona hexaloba; (K) Uvaria grandiflora; (L) Dasymaschalon trichophorum; (M) Sphaerocoryne gracilipes. — Photographs: A, Chun Chiu Pang; B–E, H–J, M, Thomas L. P. Couvreur; F, Junhao Chen; G, L, K, Xing Guo; Photos available at the World Annonaceae website64: http://annonaceae.myspecies.info/.
Skeletal representation of the best-scoring maximum likelihood tree inferred from a 754-accession dataset of eight chloroplast markers, showing topology of Malmeoideae. Tips represent genera or subdivisions when genera are not monophyletic. Higher taxon names appear at right, marked with different background colours. ML bootstrap (BS) values ≥50, MP jackknife (JK) values ≥50 and Bayesian posterior probabilities (PP) values ≥0.5 are indicated at each node: BS /JK/ PP. -, represents clade support values <50%. Numbers in brackets show numbers of sampled taxon and supplementary figures of each lineage. Tribe abbreviations: MA: Maasieae; FE: Fenerivieae; PH: Phoenicantheae; DE: Dendrokingstonieae; MO: Monocarpieae. Flower morphology of the selected genera: (A) Piptostigma multinervium; (B) Sirdavidia solannona; (C) Onychopetalum periquino; (D) Phaeanthus ophthalmicus; (E) Huberantha tanganyikensis; (F) Orophea maculata; (G) Mitrephora winitii; (H) Alphosea javanica; (I) Pseuduvaria froggattii; (J) Meiogyne heteropetala; (K) Polyalthia suberosa; (L) Stelechocarpus burahol. — Photographs: A–C, E, Thomas L. P. Couvreur; F, Junhao Chen; D, G, H, L, Xing Guo; I, J, Chun Chiu Pang; K, Daniel C. Thomas. Photos available at the World Annonaceae website64: http://annonaceae.myspecies.info/.
For the Bayesian inference (BI) analysis, the partitioned analyses based on region identity provided distinctly better explanations of the data than both analyses using the non-partitioned model and analyses of the two-partitioned dataset: 2lnB (8-partitioned over non-partitioned) = 2,493; and 2lnB (8-partitioned over 2-partitioned) = 947, significantly above the threshold value of 10. The posterior probability (PP) values derived from the analyses using eight partitions were therefore selected as the results of the Bayesian analyses.
The maximum parsimony (MP), ML, and BI analyses yielded similar topologies, differing mainly in the relative jackknife (JK), bootstrap (BS) and PP values, respectively, for particular groups (Figs 1 and 2; Supplementary Figs S1–S9). Our inferred phylogeny is generally consistent with previous phylogenetic analyses of the family, although we clarify the relationships of five genera for the first time, viz.: (1) Bocagea, sister to Trigynaea (tribe Bocageeae) (BS = 76; JK = 83; PP = 1); (2) Cardiopetalum, located in the clade together with Cymbopetalum and Porcelia (tribe Bocageeae) (BS = 100; JK = 99; PP = 1); (3) Boutiquea, sister to Neostenanthera (tribe Annoneae) (BS = 100; JK = 100; PP = 1); (4) Duckeanthus, sister to Fusaea (tribe Duguetieae) (BS = 100; JK = 100; PP = 1); and (5) Phoenicanthus, which does not belong to any currently recognised tribe, as sister to a clade comprising the tribes Dendrokingstonieae, Monocarpieae and Miliuseae (BS = 95; JK = 83; PP = 1).
The monophyletic status of the majority of the 105 sampled genera is supported in all analyses (Figs 1 and 2; Supplementary Figs S1–S9). Six genera, however, are consistently retrieved as non-monophyletic: Desmopsis (Fig. S9), Friesodielsia (Fig. S6), Klarobelia (Fig. S7), Oxandra (Fig. S7), Piptostigma (Fig. S7) and Stenanona (Fig. S9).
Discussion
Our phylogenetic reconstructions consistently retrieved 17 primary clades, which are grouped into four moderately to highly supported larger clades (Fig. 1; Supplementary Fig. S1). There is considerable congruence between these clades and the four subfamilies and 14 tribes currently recognised2. Since the phylogeny and taxonomy of these clades were discussed extensively by Chatrou et al.2 the relationships between them will not be discussed again here. We focus instead on the phylogenetic placement of the five newly sampled genera and the non-monophyletic status of several genera indicated by our large-scale phylogenetic reconstruction.
Phylogenetic relationships of previously unplaced genera
The monotypic genus Boutiquea, essentially endemic to Cameroon, is resolved within the tribe Annoneae, unambiguously supported as sister to the genus Neostenanthera, which consists of five species from West and Central Africa15 (Fig. 1; Supplementary Fig. S4). Boutiquea was already included in the tribe Annoneae but only based on pollen and morphological characters2. This sister relationship is consistent with floral and pollen morphology: both genera have an elevated floral torus, very short sepals, elongated petals, three apically connivent inner petals that form a mitriform dome over the reproductive organs16, and pollen grains with a granular infratectum that are released as tetrads17, 18. In addition, both genera have septate anthers in which the sporogenous cells are partitioned by transverse or longitudinal walls comprising sterile tissue19. Interestingly, Boutiquea platypetala (Engl. & Diels) Le Thomas was previously included in Neostenanthera, although Le Thomas20 noted that it was distinguished from other species in the genus by its sessile monocarps, which are divided into a conical apical and a hemispheric basal region by a thickened transverse ledge at the widest part, hence differing from the clearly stipitate and fusiform or ellipsoid monocarps typical of Neostenanthera 15. Given the morphological similarities between Boutiquea platypetala and Neostenanthera and the sister-group relationship between the two genera, we suggest that there is little to be gained from their continued separation. Since the generic name Neostenanthera 21 antedates that of Boutiquea 20, we recommend that the latter name should be treated as a synonym of Neostenanthera, and that the existing combination Neostenanthera platypetala (Engl. & Diels) Pellegr. be adopted.
Our phylogenetic results suggest that the Brazilian monotypic genus Duckeanthus is sister to Fusaea (BS = 100; JK = 100; PP = 1, Fig. 1; Supplementary Fig. S3) within the tribe Duguetieae; this corroborates the results of previous cladistic analyses based on morphological characters22, which indicated that Duckeanthus and Fusaea have similar inflorescences, stamen anatomy and aril structure. The two genera are also palynologically similar, with large pollen tetrads with a minutely granular exine structure18, 23, 24. Fusaea differs from Duckeanthus, however, in its fused carpels, a conspicuous ring of staminodes, and the fused calyx16.
The Neotropical genera Bocagea and Cardiopetalum are confirmed as members of the tribe Bocageeae (Fig. 1; Supplementary Fig. S1). Bocagea is shown to be sister to Trigynaea, and Cardiopetalum is retrieved in a clade together with Cymbopetalum and Porcelia. These genera, together with Hornschuchia and Mkilua, are shown to be collectively monophyletic and morphologically easily distinguished from other genera in the family by reference to their solitary internodal ebracteate pedicels that are basally articulated, and pollen that is shed in polyads comprising eight or more grains25. Apart from the African genus Mkilua, which forms the basal lineage, all other genera of this tribe are Neotropical, forming two well supported subclades, Cardiopetalum-Cymbopetalum-Porcelia (BS = 100; JK = 99; PP = 1) and Bocagea-Hornschuchia-Trigynaea (BS = 100; JK = 98; PP = 1). These inferred relationships are consistent with the previous cladogram based on morphological characters25: the Cardiopetalum-Cymbopetalum-Porcelia clade is supported by outer petals with valvate to slightly imbricate aestivation, the presence of specialised tissues on petal margins that function as pollinator food rewards, and the absence of a seed caruncle; the Bocagea-Hornschuchia-Trigynaea clade, in contrast, is united by a suite of eight characters, including the narrow floral torus, sepals that are persistent in the fruit, inner petals with a trigonous apex, few stamens, elongate anther connectives, variably tectate pollen exine, and non-articulated stigmas.
Phoenicanthus is one of the most poorly known genera in the Annonaceae, with only two species currently recognised. The genus has been placed in tribe Miliuseae in previous classifications2, 26, 27 because of their ‘miliusoid’ stamens (in which the connective does not extend over the thecae) with an obtuse apex. Miliusoid stamens have been shown to be morphologically highly variable, however, and likely homoplasious in both the Miliuseae and the wider family16, 28. The phylogenetic results presented here, however, indicate that Phoenicanthus is the next-divergent branch subsequent to the tribe Fenerivieae, and strongly supported as sister to a clade comprising the tribes Dendrokingstonieae, Monocarpieae and Miliuseae (BS = 95; JK = 83; PP = 1, Fig. 2; Supplementary Fig. S7): the tribe Miliuseae is shown as sister to the Monocarpieae, with these two tribes collectively sister to the Dendrokingstonieae. The inferred relationships within this clade clearly contradict any association of Phoenicanthus with the tribe Miliuseae.
Phoenicanthus can easily be distinguished from Fenerivia by the lack of a prominent flange immediately below the perianth, which has been interpreted as the highly reduced calyx and synapomorphic for Fenerivia 29, 30. Similarities between Phoenicanthus, Dendrokingstonia and Monocarpia include their eucamptodromous leaf venation with percurrent tertiary veins (X. Guo, pers. observ.) and very limited number of carpels (only 1–3 per flower)31. Phoenicanthus differs from the other two genera, however, by a combination of macromorphological flower and fruit characters: Dendrokingstonia and Monocarpia have considerably enlarged peltate stigmas, whorled stamens, and relatively large monocarps (3–5 cm in diameter); Phoenicanthus, in contrast, has reduced stigmas, stamens arranged in a triangular floral meristem with solitary stamens at the corners, and small monocarps (c. 1 cm in diameter).
Our molecular phylogenetic analyses indicate that Phoenicanthus cannot be accommodated in any of the existing tribes. This is supported by the morphological data, which provide strong support for distinguishing Phoenicanthus from related taxa. A new tribe is accordingly warranted, and is described below as tribe Phoenicantheae:
Phoenicantheae X. Guo & R. M. K. Saunders, tribus nov. – TYPE GENUS: Phoenicanthus Alston in Trimen, Handb. Fl. Ceylon 6: 6 (1931).
Glabrous trees; pedicel bracts present; flowers bisexual, solitary or in few-flowered fascicles, terminal; sepals 3 per flower, slightly connate; petals 6 per flower, subequal, inner petals mitriform and concave at the base; stamens 6 or 9 per flower, ‘miliusoid’ stamens (connectives not extending over thecae) with obtuse apex, triangularly arranged; carpel(s) 1–3 per flower, stigmas reduced; ovule(s) 1–2 per carpel; monocarps globose, sessile.
Comprising a single genus, Phoenicanthus, with two species, P. coriacea (Thwaites) H. Huber and P. obliquus (Hook. f. & Thomson) Alston, endemic to Sri Lanka32.
Polyphyletic and paraphyletic genera
Our phylogenetic reconstructions support the monophyletic status of the majority of genera, but six genera are not well supported as monophyletic, viz. Desmopsis, Friesodielsia, Klarobelia, Oxandra, Piptostigma and Stenanona. The large-scale phylogeny presented here provides an overview of all non-monophyletic genera, although relevant relationships have previously been reported in phylogenetic analyses targeting specific genera2, 4, 7, 11, 33,34,35.
Klarobelia (tribe Malmeeae: Fig. 2; Supplementary Fig. S7) is shown to be paraphyletic, with the monotypic genus Pseudephedranthus nested within it (BS = 83; JK = 61; PP = 1), corroborating the results of previous phylogenetic analyses2, 4. The problem is further complicated because the Klarobelia-Pseudephedranthus clade, together with Pseudomalmea, are deeply nested within Oxandra, which is itself paraphyletic (Fig. 2; Supplementary Fig. S7). The non-monophyletic status of Oxandra has been reported in previous phylogenetic studies2, 5, highlighting the problem in the current generic circumscription.
Klarobelia and Pseudomalmea species were originally classified in Malmea 36 but were subsequently removed and accommodated in two newly described genera37 on the basis of leaf, inflorescence and seed characters. In general appearance, Klarobelia and Pseudomalmea are very similar to Oxandra, although with relatively minor differences in the number of bracts per pedicel, petal shape, and monocarp stipe length38: Oxandra species have 3–6 bracts per pedicel, petals that are 4–8 mm long, and stipes shorter than 10 mm; whereas Klarobelia and Pseudomalmea species have only one or two bracts per pedicel, petals that are 7–70 mm long, and stipes longer than 10 mm. Klarobelia only differs from Pseudomalmea by its concave petals that cover the reproductive organs, which are outwardly spreading in Pseudomalmea. Pseudephedranthus also closely resembles Oxandra, with differences restricted to its longer petals and discoid stamen apices16.
One possible treatment to render Oxandra monophyletic might be to adopt a broad generic delimitation by merging Klarobelia, Pseudomalmea and Pseudephedranthus into Oxandra. Alternatively, Oxandra could be treated in a narrow sense including species located in the same clade with the type species O. lanceolata, with the remaining distantly related species transferred to other genera or segregated as a new genus. Most nodes within this clade are poorly supported (Fig. 2; Supplementary Fig. S7), however, probably due to the limited number of DNA regions sequenced (only rbcL, psbA-trnH and trnL-F are available for most species). Further studies with a more extensive taxon sampling and based on additional DNA regions are essential before validating nomenclatural changes for these genera.
The genus Piptostigma is shown to be paraphyletic, with P. fasciculatum (De Wild.) Boutique ex Fries sister to a well-supported clade (BS = 99; JK = 99; PP = 1) comprising three species of Piptostigma and four species of Polyceratocarpus (Fig. 2; Supplementary Fig. S7). These relationships are consistent with those of previous phylogenetic studies11, 35, 39. Morphological data also indicate that Piptostigma is heterogeneous, with the majority of species possessing tuberculate monocarps and sepaloid outer petals, whereas P. fasciculatum has relatively smooth monocarps and outer petals that are similar to the inner petals. Based on combined molecular and morphological data, Ghogue et al.39 have recently removed P. fasciculatum from Piptostigma, transferring it to the resurrected genus Brieya which now contains two species (Brieya fasciculata De Wild. and Brieya latipetala Exell).
The two species of Stenanona (tribe Miliuseae) sampled in the present study are retrieved in two separate lineages nested within Desmopsis, rendering the latter genus polyphyletic (Fig. 2; Supplementary Fig. S9). The relationships inferred here are partially congruent with those of a previous study35 based on two DNA regions (rbcL and trnL-F), in which two sampled species of Desmopsis formed a clade together with species of Stenanona and Stelechocarpus. Non-monophyly of Desmopsis and Stenanona was also confirmed in a recent phylogenetic study by Ortiz-Rodriguez et al.34, which was based on a more extensive taxon sampling with ten Stenanona spp. and eight Desmopsis spp. Despite the likely congeneric status of Desmopsis and Stenanona, Ortiz-Rodriguez et al. refrained from formalising any new combinations because relationships within the Desmopsis-Stenanona clade have not been fully resolved yet and several species of Desmopsis have not been formally described. Ortiz-Rodriguez et al.34 also proposed that the Neotropical clade within the Miliuseae be recognised as the subtribe Sapranthinae. Recognition of a single subtribe raises problems with the classification of the other genera in a complementary but potentially non-monophyletic subtribe: backbone relationships in the tribe are poorly resolved2, 5, 12, 29, and we believe that a subtribal classification is premature at best.
The monotypic genus Schefferomitra was recently shown to be nested within the Asian genus Friesodielsia (BS = 99; JK = 100; PP = 1), with African species that were previously placed under the latter name transferred to Afroguatteria, Monanthotaxis and Sphaerocoryne 7. Detailed examination of the morphological characters of the two lineages supported the phylogenetic relationship and suggested that there are no convincing criteria to support the continued recognition of Friesodielsia and Schefferomitra as distinct genera. Although the latter name has nomenclatural priority, Guo et al.40 proposed conservation of the name Friesodielsia in order to promote nomenclatural stability; no formal nomenclatural change has been made, however, pending the decision by the Nomenclature Committee for Vascular Plants. In addition to addressing the polyphyletic status of Friesodielsia, Guo et al.7 also amended the generic delimitation of Monanthotaxis by including the former genera Exellia and Gilbertiella.
Intraspecific non-monophyly in Guatteria
Guatteria is a large Neotropical genus with 177 species currently recognised41. The phylogenetic relationships within the genus retrieved here (Fig. S2) are largely congruent with those published previously42. Multiple accessions of more than 10 species do not form a well-supported clade, however. The non-monophyly of some species (e.g. G. amplifolia, G. hirsuta and G. punctata) is strongly supported: two accessions of G. amplifolia, for example, are retrieved as sister to G. latifolia with strong Bayesian support, whilst a third accession is shown to be more closely related to G. jefensis (BS = 86; JK = 85; PP = 1). The non-monophyly of these species may be the result of misidentification as they belong to several species complexes with problematic species delimitations41.
Other conflicts lack statistical support, however. The few cpDNA regions used (only matK, psbA-trnH, rbcL and trnL-F have been sequenced for the majority of species) contain limited phylogenetic information, resulting in poor resolution in this part of the tree. Additional unlinked data from different genomes are necessary to improve resolution, identifying potential gene tree incongruence and differentiate likely underlying biological causes such as incomplete lineage sorting, introgression, and/or unrecognised paralogy.
Genera not sampled
Although we initially planned to achieve a comprehensive sampling of all genera in the Annonaceae, PCR reactions of Froesiodendron were unsuccessful due to the poor quality of available leaf material. Froesiodendron comprises three species from tropical South America, and has been inferred to belong to the tribe Bocageeae based on its solitary internodal ebracteate pedicels, septate stamens and pollen shed as polyads2, 16, 25. Morphological cladistic analyses25 furthermore suggest that Froesiodendron is more closely related to Cardiopetalum, Cymbopetalum and Porcelia than to other genera in the tribe. These four genera are united by outer petals that show valvate to slightly imbricate aestivation and with specialised beetle-feeding tissues on the petal margins.
A molecular phylogenetic re-evaluation of the circumscription of Uvaria 43 led to several satellite genera being subsumed and many species transferred to an expanded Uvaria. Although all Australian representatives of Melodorum were transferred to Uvaria in this process, 11 species remained; the generic name Melodorum was not synonymised with Uvaria due to typification problems, with Melodorum long confused with Sphaerocoryne: the former name has been incorrectly applied to species belonging to the latter27, 44. It seems likely that the name of the type species, M. fruticosum Lour., has been widely misapplied in many published phylogenies and that the specimen used may represent a species of Sphaerocoryne 43. Further molecular and morphological studies with more extensive taxon sampling and a re-evaluation of nomenclatural type specimens are required to clarify the nomenclatural problems associated with the application of the name Melodorum.
Supermatrix and large-scale phylogenetic reconstruction
Results from our supermatrix analyses are promising. Even though the concatenated matrix had c. 55% missing data, we found that the generic and tribal positions of most species were consistent with previous taxonomic research, and often very strongly supported. We compared the results of our tree searches with those from the next-largest available phylogeny for the family (based on a 193-species dataset)2, with nodes excluded within the poorly resolved tribe Miliuseae (Supplementary Table S1). Of the total 25 major nodes that differed between two analyses, all but three of the nodes of our analyses are better resolved and/or have better support values than in the previous 193-species phylogeny2. This improved phylogenetic performance suggests that the strategy employed in this study of maximising the number of loci and taxa has greater power for resolving relationships, particularly at deeper nodes in the phylogeny, than traditional approaches in which only one or two species of each genus are included as placeholders, despite highly incomplete alignment.
Supermatrix methods offer a variety of advantages, including the ability to reconstruct more inclusive phylogenies at broad scales with minimal investment in sequencing45, 46. These methods present their own challenges, however, including issues of sparse alignment, data integrity, computational power and time efficiency47. Our sampling criterion has largely overcome these problems, however, and greatly facilitated the integration of the different DNA regions used in this study. Specifically, we excluded accessions with data for fewer than three regions available. In the concatenated dataset, 96% accessions have trnL-F sequence data, 86% have matK data, and 84% have rbcL data. Thus, most species have comparable data for at least three regions, which may have greatly facilitated tree reconstruction despite lacking other regions. Importantly, our study also provides a framework to which additional sequences can readily be added in future research. We anticipate that this large-scale phylogeny will be of broad utility for many areas of Annonaceae research, including historical biogeography, diversification rate studies and ecology.
Materials and Methods
Taxon and DNA region sampling
We adopted a supermatrix approach, integrating available data for 858 Annonaceae accessions downloaded from the nucleotide database of National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Drepananthus longiflorus, Hexalobus monopetalus, Melodorum fruticosum, Onychopetalum amazonicum, Unonopsis elegantissima, Unonopsis perrottetii and Unonopsis rufescens were excluded due to ambiguous identifications, and Monoon borneense, Pseudoxandra bahiensis and Pseudoxandra cuspidata were excluded due to hard incongruence between DNA markers. An additional 107 accessions were excluded for one or a combination of the following reasons: (1) multiple accessions of a single species (except for Guatteria which shows problems of intraspecific non-monophyly); (2) specimen only identified to genus level; or (3) data for fewer than three DNA regions available. Exceptions were made regarding the latter criterion for certain accessions if genera were represented by fewer than three accessions in total. Additionally, data for eight accessions representing five genera (Bocagea, Boutiquea, Cardiopetalum, Duckeanthus and Phoenicanthus) were newly generated, with voucher information provided in Supplementary Appendix I. Species belonging to three other families in the Magnoliales, including the Myristicaceae (Myristica fragrans and Coelocaryon preussii), Magnoliaceae (Magnolia kobus and Liriodendron chinense) and Eupomatiaceae (Eupomatia bennettii), were selected as outgroups. The final matrix therefore comprised 749 ingroup and five outgroup accessions (see Appendix I for voucher information), representing c. 98% of generic diversity and c. 29% of species diversity in the Annonaceae.
DNA sequences of eight chloroplast regions (rbcL, matK, ndhF, psbA-trnH, trnL-F, atpB-rbcL, trnS-G and ycf1), which are commonly used in Annonaceae phylogenetics, were downloaded from the nucleotide database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) or generated for the newly added samples in this study. Attempts at sequencing two genera, Melodorum and Froesiodendron, were unsuccessful due to the poor quality of available leaf materials. GenBank accession number for all samples included in the analyses are given in Supplementary Appendix II.
DNA extraction, amplification and sequencing
Total DNA was isolated from herbarium material using the innuPrep Plant DNA Kit (Analytik Jena, Jena, Germany) following the manufacturer’s instructions. Polymerase chain reaction (PCR) amplification and sequencing were performed using the same procedures as previously described8, 48.
Sequence assembly, alignment and phylogenetic analyses
Sequence fragments were edited and assembled using GeneiousPro v.7.1.9 (Biomatters; http://www.geneious.com). Sequences of individual regions were subsequently aligned automatically using the MAFFT plugin49 in Geneious with default settings, and then manually edited and optimised. Characters in regions for which alignment was ambiguous or included inversions and short repetitive sequences were excluded from the analyses.
Phylogenetic reconstruction was performed using maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI) methods. DNA sequences for the individual DNA regions were concatenated rather than analysed independently as chloroplast DNA is inherited as a unit.
For the MP analyses, all characters were treated as independent and of equal weight, with gaps treated as missing data. A heuristic search was performed in PAUP* v.4.0b1050 with 2,000 random addition sequence replicates with TBR branch-swapping, saving 10 trees per replicate. The most parsimonious trees were summarised using a strict consensus tree. The robustness of the phylogenetic relationships was evaluated using the jackknife (JK) method51 with the removal probability set to approximately e−1 (36.7879%), and “jac” resampling emulated. 1,000 JK replicates were performed with 100 random addition tree bisection-reconnection searches (each with a maximum of 10 trees held) per replicate.
ML analyses were performed using RAxML v.8.2.652 provided by the CIPRES Science Gateway53. The dataset was separated into eight partitions based on DNA region identity. 1,000 analyses were run from distinct random stepwise addition sequence MP starting trees under the general time-reversible nucleotide substitution model with among-site rate variation modelled with a gamma distribution (GTR + Γ). Bootstrap support (BS) values were used to estimate clade support, based on 1,000 non-parametric bootstrap replicates.
BI analysis was undertaken using MrBayes v.3.2.654 with three distinct partitioning strategies: (1) non-partitioned; (2) 2-partitioned, distinguishing coding (matK, ndhF, rbcL and ycf1) and non-coding (atpB-rbcL, psbA-trnH, trnL-F and trnS-G) regions; and (3) 8-partitioned, according to DNA region identity. The appropriate DNA substitution model for each locus and concatenated matrix was determined using MrModeltest v.2.355, applying the Akaike Information Criterion. For the 2-partitioned and 8-partitioned analyses, the parameter values (NST and gamma distributed rates) for each partition were allowed to evolve independently using the unlinked setting. Four Markov chain Monte Carlo (MCMC) chains were run, each beginning with a random tree and sampling one tree every 1,000 generations for 20 million generations. The mean branch length prior was reset from the default mean (0.1) to 0.01 (brlenspr = unconstrained: exponential (100.0)) to reduce the likelihood of stochastic entrapment in local tree length optima56, 57. Convergence was assessed using the standard deviation of split frequencies, with values <0.01 interpreted as indicating good convergence. The first 25% of samples (5,000 trees) were discarded as burn-in, and the post-burn-in samples summarised as a 50% majority-rule consensus tree.
Stationarity, convergence and a suitable effective sample size were assessed using Tracer v.1.558 and were visually checked using the Cumulative and Compare functions in AWTY59. Inference of the analyses using the three partitioning schemes was assessed with Bayes factor comparison. The best-performing partitioning strategy was selected by applying the criterion of 2ln Bayes factor >10 as strong evidence in favour of a particular model60, 61.
Bootstrap/jackknife values of 50–74% were considered as weak support, 75–84% as moderate support, and 85–100% as strong support. For BI, the estimation of branch support accompanies the tree estimation and is reflected by posterior probabilities (PP)62; branches with PP values ≥0.95 are considered well supported, and <0.95 not supported63.
Data availability
The data used in this study are available for download from the nucleotide database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). See Supplementary Appendix II for the GenBank accession numbers of all samples included in the analyses.
References
Massoni, J., Forest, F. & Sauquet, H. Increased sampling of both genes and taxa improves resolution of phylogenetic relationships within Magnoliidae, a large and early-diverging clade of angiosperms. Molec. Phylogen. Evol. 70, 84–93 (2014).
Chatrou, L. W. et al. A new subfamilial and tribal classification of the pantropical flowering plant family Annonaceae informed by molecular phylogenetics. Bot. J. Linn. Soc. 169, 5–40 (2012).
Richardson, J. E., Chatrou, L. W., Mols, J. B., Erkens, R. H. J. & Pirie, M. D. Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Philos. Trans. Roy Soc. Lond., ser. B 359, 1495–1508 (2004).
Pirie, M. D., Chatrou, L. W., Mols, J. B., Erkens, R. H. J. & Oosterhof, J. ‘Andean-centred’ genera in the short-branch clade of Annonaceae: Testing biogeographical hypotheses using phylogeny reconstruction and molecular dating. J. Biogeogr. 33, 31–46 (2006).
Couvreur, T. L. P. et al. Early evolutionary history of the flowering plant family Annonaceae: Steady diversification and boreotropical geodispersal. J. Biogeogr. 38, 664–680 (2011).
Li, P. S., Thomas, D. C. & Saunders, R. M. K. Phylogenetic reconstruction, morphological diversification and generic delimitation of Disepalum (Annonaceae). PloS One 10, e0143481 (2015).
Guo, X. et al. Cutting up the climbers: evidence for extensive polyphyly in Friesodielsia (Annonaceae) necessitates generic realignment across the tribe Uvarieae. Taxon 66, 3–19 (2017).
Tang, C. C., Thomas, D. C. & Saunders, R. M. K. Molecular phylogenetics of the species-rich angiosperm genus Goniothalamus (Annonaceae) inferred from nine chloroplast DNA regions: Synapomorphies and putative correlated evolutionary changes in fruit and seed morphology. Molec. Phylogen. Evol. 92, 124–139 (2015).
Chaowasku, T., Johnson, D. M., van der Ham, R. W. J. M. & Chatrou, L. W. Characterization of Hubera (Annonaceae), a new genus segregated from Polyalthia and allied to Miliusa. Phytotaxa 69, 33–56 (2012).
Xue, B., Thomas, D. C., Chaowasku, T., Johnson, D. M. & Saunders, R. M. K. Molecular phylogenetic support for the taxonomic merger of Fitzalania and Meiogyne (Annonaceae): new nomenclatural combinations under the conserved name. Meiogyne. Syst. Bot. 39, 396–404 (2014).
Couvreur, T. L. P., Niangadouma, R., Sonké, B. & Sauquet, H. Sirdavidia, an extraordinary new genus of Annonaceae from Gabon. PhytoKeys 46, 1–19 (2015).
Xue, B., Su, Y. C. F., Thomas, D. C. & Saunders, R. M. K. Pruning the polyphyletic genus Polyalthia (Annonaceae) and resurrecting the genus Monoon. Taxon 61, 1021–1039 (2012).
Guo, X. et al. Reassessing the taxonomic status of two enigmatic Desmos species (Annonaceae): morphological and molecular phylogenetic support for a new genus. Wangia. J. Syst. Evol. 52, 1–15 (2014).
Chaowasku, T., van der Ham, R. W. J. M. & Chatrou, L. W. Integrative systematics supports the establishment of Winitia, a new genus of Annonaceae (Malmeoideae, Miliuseae) allied to Stelechocarpus and Sageraea. Syst. Biodivers. 11, 195–207 (2013).
Fero, M., Aedo, C., Cabezas, F. & Velayos, M. Taxonomic Revision of Neostenanthera (Annonaceae). Syst. Bot. 39, 17–30 (2014).
van Heusden, E. C. H. Flowers of Annonaceae: Morphology, classification, and evolution. Blumea Suppl. 7, 1–218 (1992).
Le Thomas, A. Ultrastructural characters of the pollen grains of African Annonaceae and their significance for the phylogeny of primitive angiosperms: Part I. Pollen Spores 22, 267–342 (1980).
Doyle, J. A. & Le Thomas, A. Evolution and phylogenetic significance of pollen in Annonaceae. Bot. J. Linn. Soc. 169, 190–221 (2012).
Tsou, C. H. & Johnson, D. M. Comparative development of aseptate and septate anthers of Annonaceae. Amer. J. Bot. 90, 832–848 (2003).
Le Thomas, A. Un nouveau genre africain d’Annonacées: Boutiquea Le Thomas. Adansonia 5, 531–535 (1965).
Exell, A. W. Mr John Gossweiler’s plants from Angola and Portuguese Congo. J. Bot., Brit. For. 73, 2–7 (1935).
Chatrou, L. W., Koek-Noorman, J. & Maas, P. J. M. Studies in Annonaceae XXXVI. The Duguetia alliance: where the ways part. Ann. Missouri Bot. Gard. 87, 234–245 (2000).
Walker, J. W. Pollen morphology, phytogeography, and phylogeny of the Annonaceae. Contr. Gray Herb. 202, 1–131 (1971).
Le Thomas, A., Lugardon, B. & Doyle, J. A. Pollen ultrastructure and relationships of Fusaea (Baillon) Safford and Duguetia A. Saint-Hilaire (Annonaceae). Rev. Palaeobot. Palynol. 83, 55–64 (1994).
Johnson, D. M. & Murray, N. A. Synopsis of the tribe Bocageeae (Annonaceae), with revisions of Cardiopetalum, Froesiodendron, Trigynaea, Bocagea, and Hornschuchia. Brittonia 47, 248–319 (1995).
Hooker, J. D. & Thomson, T. In The Flora of British India, Vol. I (ed. Hooker, J. D.) Annonaceae. 45–94 (L. Reeve, 1872).
Kessler, P. J. A. In The Families and Genera of Vascular Plants (eds. Kubitzki, K., Rohwer, J. G. & Bittrich, V.), Annonaceae. 93–129 (Springer, 1993).
Chaowasku, T. et al. A plastid DNA phylogeny of tribe Miliuseae: insights into relationships and character evolution in one of the most recalcitrant major clades of Annonaceae. Am. J. Bot. 101, 691–709 (2014).
Saunders, R. M. K., Su, Y. C. F. & Xue, B. Phylogenetic affinities of Polyalthia species (Annonaceae) with columellar-sulcate pollen: enlarging the Madagascan endemic genus Fenerivia. Taxon 60, 1407–1416 (2011).
Xue, B. & Saunders, R. M. K. Reassessing morphological homologies in the early-divergent angiosperm Fenerivia (Annonaceae) based on floral vascular anatomy: significance for interpreting putative homeotic mutations. PLoS One 8, e81923 (2013).
Chaowasku, T., Kessler, P. J. A. & van der Ham, R. W. J. M. A taxonomic revision and pollen morphology of the genus Dendrokingstonia (Annonaceae). Bot. J. Linn. Soc. 168, 76–90 (2012).
Huber, H. In A Revised Handbook to the Flora of Ceylon, Vol. V (ed. Dassanayake, M. D.) Annonaceae. 1–75 (Amerind, 1985).
Mols, J. B. et al. Identifying clades in Asian Annonaceae: monophyletic genera in the polyphyletic Miliuseae. Amer. J. Bot. 91, 590–600 (2004).
Ortiz-Rodriguez, A. E., Ruiz-Sanchez, E. & Ornelas, J. F. Phylogenetic relationships among members of the Neotropical clade of Miliuseae (Annonaceae): generic non-monophyly of Desmopsis and Stenanona. Syst. Bot. 41, 815–822 (2016).
Couvreur, T. L., van der Ham, R. W. J. M., Mbele, Y. M., Mbago, F. M. & Johnson, D. M. Molecular and morphological characterization of a new monotypic genus of Annonaceae, Mwasumbia from Tanzania. Syst. Bot. 34, 266–276 (2009).
Fries, R. E. Revision der Arten einiger Annonaceen-Gattungen IV. Acta Horti Berg. 12, 221–288 (1937).
Chatrou, L. W. In Changing genera: Systematic studies in Neotropical and West African Annonaceae (ed. Chatrou, L. W.), 105–192. Revision of the Malmea alliance: Malmea and three new, Neotropical genera: Klarobelia, Mosannona, and Pseudomalmea. (University of Utrecht, 1998).
Couvreur, T. L. P., Maas, P. J. M., Meinke, S., Johnson, D. M. & Keßler, P. J. A. Keys to the genera of Annonaceae. Bot. J. Linn. Soc. 169, 74–83 (2012).
Ghogue, J.-P., Sonké, B. & Couvreur, T. L. P. Taxonomic revision of the African genera Brieya and Piptostigma (Annonaceae). Plant Ecology and Evolution 150, 173–216 (2017). DOI: 10.5091/plecevo.2017.1137
Guo, X., Tang, C. C. & Saunders, R. M. K. Proposal to conserve the name Friesodielsia against Schefferomitra (Annonaceae). Taxon 66, 204–205 (2017).
Maas, P. J. M. et al. Confronting a morphological nightmare: revision of the Neotropical genus Guatteria (Annonaceae). Blumea 60, 1–219 (2015).
Erkens, R. H., Chatrou, L. W., Koek-Noorman, J., Maas, J. W. & Maas, P. J. Classification of a large and widespread genus of Neotropical trees, Guatteria (Annonaceae) and its three satellite genera Guatteriella, Guatteriopsis and Heteropetalum. Taxon 56, 757–774 (2007).
Zhou, L., Su, Y. C. F., Chalermglin, P. & Saunders, R. M. K. Molecular phylogenetics of Uvaria (Annonaceae): relationships with Balonga, Dasoclema and Australian species of Melodorum. Bot. J. Linn. Soc. 163, 33–43 (2010).
Sinclair, J. A revision of the Malayan Annonaceae. Gard. Bull. Singapore 14, 368–371 (1955).
Davis, B. W., Li, G. & Murphy, W. J. Supermatrix and species tree methods resolve phylogenetic relationships within the big cats, Panthera (Carnivora: Felidae). Molec. Phylogen. Evol. 56, 64–76 (2010).
Wolsan, M. & Sato, J. J. Effects of data incompleteness on the relative performance of parsimony and Bayesian approaches in a supermatrix phylogenetic reconstruction of Mustelidae and Procyonidae (Carnivora). Cladistics 26, 168–194 (2010).
Hinchliff, C. E. & Roalson, E. H. Using supermatrices for phylogenetic inquiry: an example using the sedges. Syst. Biol. 62, 205–219 (2013).
Tang, C. C., Thomas, D. C. & Saunders, R. M. K. Molecular and morphological data supporting phylogenetic reconstruction of the genus Goniothalamus (Annonaceae), including interpretations of previous infrageneric classifications. Data Brief 4, 410–421 (2015).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acids Res. 30, 3059–3066 (2002).
Swofford, D.L. PAUP*: Phylogenetic analysis using parsimony (*and other methods), ver. 4.0b10. (Sinauer, 2002).
Farris, J. S., Albert, V. A., Källersjö, M., Lipscomb, D. & Kluge, A. G. Parsimony jackknifing outperforms neighbor-joining. Cladistics 12, 99–124 (1996).
Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE). 1–8 (New Orleans: IEEE. 2010).
Ronquist, F. et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Nylander, J. A. A. MrModeltest, version 2 [online]. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. Available from http://www.abc.se/~nylander/mrmodeltest2/mrmodeltest2.html. (Accessed: 1st September 2016) (2004).
Brown, J. M., Hedtke, S. M., Lemmon, A. R. & Lemmon, E. M. When trees grow too long: Investigating the causes of highly inaccurate Bayesian branch-length estimates. Syst. Biol. 59, 145–161 (2010).
Marshall, D. C. Cryptic failure of partitioned Bayesian phylogenetic analyses: lost in the land of long trees. Syst. Biol. 59, 108–117 (2010).
Rambaut, A. & Drummond, A.J. Tracer, version 1.5. Available from http://beast.bio.ed.ac.uk/Tracer. (Accessed: 1st September 2016) (2009).
Nylander, J. A. A., Wilgenbusch, J. C., Warren, D. L. & Swofford, D. L. AWTY (Are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583 (2008).
Kass, R. E. & Raftery, A. E. Bayes factors. J. Amer. Statist. Assoc. 90, 773–795 (1995).
Nylander, J. A. A., Ronquist, F., Huelsenbeck, J. P. & Nieves-Aldrey, J. L. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67 (2004).
Larget, B. & Simon, D. L. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molec. Biol. Evol. 16, 750–759 (1999).
Yang, Z. & Rannala, B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Molec. Biol. Evol. 14, 717–724 (1997).
Couvreur T. L. P. & Annonaceae community Available at: http://annonaceae.myspecies.info/. (Accessed: 1st September 2016) (2014).
Junikka, L., Maas, P. J. M., Maas-van de Kamer, H. & Westra, L. T. Revision of Oxandra (Annonaceae). Blumea 61, 215–266 (2016).
Couvreur, T. L. P. Revision of the African genus Uvariastrum (Annonaceae). PhytoKeys 33, 1–40 (2014).
Couvreur, T. L. P. & Niangadouma, R. New species of Uvariopsis (Annonaceae) and Laccosperma (Arecaceae/Palmae) from Monts de Cristal, Gabon. PhytoKeys 68, 1–8 (2016).
Turner, I. M. & Utteridge, T. M. A. A new species and a new combination in Meiogyne (Annonaceae) of New Guinea. Contributions to the Flora of Mt Jaya, XXI. Kew Bull. 70, 1–5 (2015).
Rajkumar, G., Alister, M., Nazarudeen, A. & Pandurangan, A. G. Miliusa sahyadrica, a new species of Annonaceae from the Western Ghats, India. Phytotaxa 284, 211–217 (2016).
Chaowasku, T., Kessler, P. J., Punnadee, S. & Van der Ham, R. W. Taxonomic novelties and pollen morphological study in the genus Neo-uvaria (Annonaceae). Phytotaxa 32, 27–42 (2011).
Ortiz-Rodriguez, A. E., Escobar-Castellanos, M. A. & Pérez-Farrera, M. A. Phylogenetic analyses and morphological characteristics support the description of a second species of Tridimeris (Annonaceae). PhytoKeys 74, 79–96 (2016).
Xue, B., Tan, Y. H. & Ye, X. E. The identity of Polyalthia florulenta (Annonaceae): a second species of Wangia in China. Phytotaxa 283, 163–171 (2016).
Acknowledgements
This research was supported by a grant from the University of Hong Kong Research Committee (awarded to CCT and RMKS). We are grateful to the curators of BR, E, L, NY, P and WAG herbaria for providing leaf material; Junhao Chen and Chun Chiu Pang for providing photographs; Roy Erkens for pre-submission feedback; and Laura Wong for general technical assistance.
Author information
Authors and Affiliations
Contributions
Designed the project: R.M.K.S. and C.C.T. Collected materials and generated data: C.C.T. (with two DNA regions of Boutiquea platypetala provided by T.L.P.C.). Analysed the data: X.G. Wrote the paper: X.G., R.M.K.S., D.C.T. and T.L.P.C.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, X., Tang, C.C., Thomas, D.C. et al. A mega-phylogeny of the Annonaceae: taxonomic placement of five enigmatic genera and support for a new tribe, Phoenicantheae. Sci Rep 7, 7323 (2017). https://doi.org/10.1038/s41598-017-07252-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07252-2
This article is cited by
-
Discovery and conservation of Monanthotaxis bali (Annonaceae) a new Critically Endangered (possibly extinct) montane forest treelet from Bali Ngemba, North West Region, Cameroon
Kew Bulletin (2023)
-
Lukea gen. nov. (Monodoreae-Annonaceae) with two new threatened species of shrub from the forests of the Udzungwas, Tanzania and Kaya Ribe, Kenya
Kew Bulletin (2022)
-
Larvicidal effect from different Annonaceae species on Culex quinquefasciatus
Environmental Science and Pollution Research (2020)
-
Formation pattern in five types of pollen tetrad in Pseuduvaria trimera (Annonaceae)
Protoplasma (2019)
-
Revision and new species of the African genus Mischogyne (Annonaceae)
Kew Bulletin (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.