Abstract
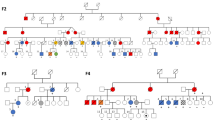
Although somatic mutations of DNA repair genes are frequent in mantle cell lymphoma (MCL), our understanding of their germline defects is limited. In a Chinese family with maternal Lynch syndrome and paternal B cell non-Hodgkin lymphoma, one sibling developed both Lynch syndrome and MCL. Lynch syndrome is caused by heterozygous mutations in mismatch repair (MMR) genes. To understand the genetic predispositions in the family, we performed exome sequencing and analyses of affected individuals and their tumor samples. A novel germline indel, MLH1 Gly101fsX1, was identified as the cause of Lynch syndrome, and unstable microsatellite loci and mutational signatures as evidence of defective MMR were revealed in the MCL sample. Furthermore, we included additional 15 MCL patients with early onset, and found by exome sequencing that 11 patients carried heterozygous germline variants of 20 DNA repair genes, including MSH2 in MMR. In the MCL with MSH2 Arg359fsX16, unstable microsatellite loci and defective MMR signatures were also found. In addition, five patients also had heterozygous germline variants of genes involved in B cell functions. Thus, our study found germline variants of genes in single-strand break repair, double-strand break repair, and Fanconi anemia pathway in early onset MCL; and for the first time we identified germline defects of MMR in two MCLs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dreyling M, Campo E, Hermine O, Jerkeman M, Le Gouill S, Rule S. et al. Newly diagnosed and relapsed mantle cell lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(Suppl 4):iv62–71.
Vogt N, Dai B, Erdmann T, Berdel WE, Lenz G. The molecular pathogenesis of mantle cell lymphoma. Leuk Lymphoma. 2017;58:1530–7.
Cheah CY, Seymour JF, Wang ML. Mantle cell lymphoma. J Clin Oncol. 2016;34:1256–69.
Puente XS, Jares P, Campo E. Chronic lymphocytic leukemia and mantle cell lymphoma: crossroads of genetic and microenvironment interactions. Blood. 2018;131:2283–96.
Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–62.
Perez-Galan P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood. 2011;117:26–38.
Sud A, Chattopadhyay S, Thomsen H, Sundquist K, Sundquist J, Houlston RS, et al. Analysis of 153 115 patients with hematological malignancies refines the spectrum of familial risk. Blood. 2019;134:960–9.
Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–94.
Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–4.
Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Ferrari V, Broggini M, Baratelli E, Galvani L, Colombo C. [Association of Lynch syndrome and non-Hodgkin’s malignant lymphoma. A case report]. Recenti Prog Med. 1999;90:81–3.
Rosty C, Briere J, Cellier C, Delabesse E, Carnot F, Barbier JP. et al.Association of a duodenal follicular lymphoma and hereditary nonpolyposis colorectal cancer.Mod Pathol.2000;13:586–90.
Teruya-Feldstein J, Greene J, Cohen L, Popplewell L, Ellis NA, Offit K. Analysis of mismatch repair defects in the familial occurrence of lymphoma and colorectal cancer. Leuk Lymphoma. 2002;43:1619–26.
Hirano K, Yamashita K, Yamashita N, Nakatsumi Y, Esumi H, Kawashima A, et al. Non-Hodgkin’s lymphoma in a patient with probable hereditary nonpolyposis colon cancer: report of a case and review of the literature. Dis Colon Rectum. 2002;45:273–9.
Cheah CY, Dsouza L, Taggart MW, Schlette EJ, Turturro F. Diffuse large B-cell lymphoma with microsatellite instability developing in the setting of Muir-Torre variant hereditary non-polyposis colon cancer. J Clin Pathol. 2015;68:755–7.
Pineda M, Castellsague E, Musulen E, Llort G, Frebourg T, Baert-Desurmont S, et al. Non-Hodgkin lymphoma related to hereditary nonpolyposis colorectal cancer in a patient with a novel heterozygous complex deletion in the MSH2 gene. Genes Chromosomes Cancer. 2008;47:326–32.
Kratz CP, Niemeyer CM, Juttner E, Kartal M, Weninger A, Schmitt-Graeff A, et al. Childhood T-cell non-Hodgkin’s lymphoma, colorectal carcinoma and brain tumor in association with cafe-au-lait spots caused by a novel homozygous PMS2 mutation. Leukemia. 2008;22:1078–80.
Kruger S, Kinzel M, Walldorf C, Gottschling S, Bier A, Tinschert S, et al. Homozygous PMS2 germline mutations in two families with early-onset haematological malignancy, brain tumours, HNPCC-associated tumours, and signs of neurofibromatosis type 1. Eur J Hum Genet. 2008;16:62–72.
Ripperger T, Beger C, Rahner N, Sykora KW, Bockmeyer CL, Lehmann U, et al. Constitutional mismatch repair deficiency and childhood leukemia/lymphoma-report on a novel biallelic MSH6 mutation. Haematologica. 2010;95:841–4.
Ripperger T, Schlegelberger B. Acute lymphoblastic leukemia and lymphoma in the context of constitutional mismatch repair deficiency syndrome. Eur J Med Genet. 2016;59:133–42.
Kotoula V, Hytiroglou P, Kaloutsi V, Barbanis S, Kouidou S, Papadimitriou CS. Mismatch repair gene expression in malignant lymphoproliferative disorders of B-cell origin. Leuk Lymphoma. 2002;43:393–9.
Sanz-Vaque L, Colomer D, Bosch F, Lopez-Guillermo A, Dreyling MH, Bosch F, et al. Microsatellite instability analysis in typical and progressed mantle cell lymphoma and B-cell chronic lymphocytic leukemia. Haematologica. 2001;86:181–6.
Renkonen E, Zhang Y, Lohi H, Salovaara R, Abdel-Rahman WM, Nilbert M, et al. Altered expression of MLH1, MSH2, and MSH6 in predisposition to hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2003;21:3629–37.
Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–9.
Du Z, Ma L, Qu H, Chen W, Zhang B, Lu X, et al. Whole genome analyses of chinese population and de novo assembly of A Northern Han Genome. Genomics Proteom Bioinforma. 2019;17:229–47.
Tian T, Li J, Xue T, Yu B, Li X, Zhou X. Microsatellite instability and its associations with the clinicopathologic characteristics of diffuse large B-cell lymphoma. Cancer Med. 2020;9:2330–42.
Cuceu C, Colicchio B, Jeandidier E, Junker S, Plassa F, Shim G. et al. Independent mechanisms lead to genomic instability in hodgkin lymphoma: microsatellite or chromosomal instability (dagger). Cancers. 2018;10:233.
de Miranda NF, Peng R, Georgiou K, Wu C, Falk Sorqvist E, Berglund M, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med. 2013;210:1729–42.
Degroote A, Knippenberg L, Vander Borght S, Spaepen M, Matthijs G, Schaeffer DF, et al. Analysis of microsatellite instability in gastric mucosa-associated lymphoid tissue lymphoma. Leuk Lymphoma. 2013;54:812–8.
Smedby KE, Sampson JN, Turner JJ, Slager SL, Maynadie M, Roman E, et al. Medical history, lifestyle, family history, and occupational risk factors for mantle cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:76–86.
Moffitt AB, Dave SS. Clinical applications of the genomic landscape of aggressive Non-Hodgkin lymphoma. J Clin Oncol. 2017;35:955–62.
Taylor J, Xiao W, Abdel-Wahab O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood. 2017;130:410–23.
Elenitoba-Johnson KSJ, Lim MS. New insights into lymphoma pathogenesis. Annu Rev Pathol. 2018;13:193–217.
Huet S, Sujobert P, Salles G. From genetics to the clinic: a translational perspective on follicular lymphoma. Nat Rev Cancer. 2018;18:224–39.
Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21:1199–208.
Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21:1190–8.
Lindsley AW, Saal HM, Burrow TA, Hopkin RJ, Shchelochkov O, Khandelwal P, et al. Defects of B-cell terminal differentiation in patients with type-1 Kabuki syndrome. J Allergy Clin Immunol. 2016;137:179–87. e10.
Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15:334–46.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Adzhubei I, Jordan DM, Sunyaev SR Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013; Chapter 7:Unit7 20.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9.
Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7.
Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15.
Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92.
Acknowledgements
This work was supported by grants from the Key Research Program of the Chinese Academy of Sciences (KJZD-EW-L06-2) (CZ), the National Natural Science Foundation of China (81972807, 81670187 and 81470368) (YS and JZ), the National High Technology Research and Development Program (863 Program) of China (2012AA022502) (CZ), the Beijing Natural Science Foundation (7202025) (YS), and the China Postdoctoral Science Foundation (2013M541009) (XW). We thank Sheng Li for the help in the interview of relatives of the family and collection of their samples. We are grateful to Yunfei Shi for pathological review. Further thanks are due to Wen Zheng, Ningjing Lin, Meifeng Tu, Xiaopei Wang, Lingyan Ping, Zhitao Ying, Lijuan Deng, Chen Zhang, Tingting Du, Meng Wu, Xin Len, and Feier Feng for their help in collection of clinical information. We are grateful to Caixia Guo for advice and help in revision.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, X., Song, Y., Chen, W. et al. Germline variants of DNA repair genes in early onset mantle cell lymphoma. Oncogene 40, 551–563 (2021). https://doi.org/10.1038/s41388-020-01542-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01542-2
This article is cited by
-
Second primary malignancies in non-Hodgkin lymphoma: epidemiology and risk factors
Annals of Hematology (2023)
-
Germline CHEK2 and ATM Variants in Myeloid and Other Hematopoietic Malignancies
Current Hematologic Malignancy Reports (2022)