Abstract
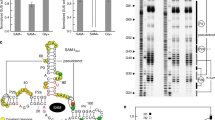
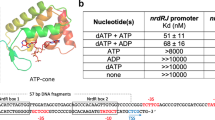
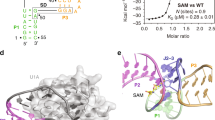
Riboswitches are metabolite-binding RNA structures that serve as genetic control elements for certain messenger RNAs. These RNA switches have been identified in all three kingdoms of life and are typically responsible for the control of genes whose protein products are involved in the biosynthesis, transport or utilization of the target metabolite. Herein, we report that a highly conserved RNA domain found in bacteria serves as a riboswitch that responds to the coenzyme S-adenosylmethionine (SAM) with remarkably high affinity and specificity. SAM riboswitches undergo structural reorganization upon introduction of SAM, and these allosteric changes regulate the expression of 26 genes in Bacillus subtilis. This and related findings indicate that direct interaction between small metabolites and allosteric mRNAs is an important and widespread form of genetic regulation in bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nahvi, A. et al. Genetic control by a metabolite binding mRNA. Chem. Biol. 9, 1043–1049 (2002).
Winkler, W., Nahvi, A. & Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Winkler, W.C., Cohen-Chalamish, S. & Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 99, 15908–15913 (2002).
Mironov, A.S. et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756 (2002).
Sudarsan, N., Barrick, J.E. & Breaker, R.R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 644–647 (2003).
Gelfand, M.S., Mironov, A.A., Jomantas, J., Kozlov, Y.I. & Perumov, D.A. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15, 439–442 (1999).
Miranda-Rios, J., Navarro, M. & Soberón, M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. USA 98, 9736–9741 (2001).
Stormo, G.D. & Ji, Y. Do mRNAs act as direct sensors of small molecules to control their expression? Proc. Natl. Acad. Sci. USA 98, 9465–9467 (2001).
Rodionov, D.A., Vitreschak, A.G., Mironov, A.A. & Gelfand, M.S. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 277, 48949–48959 (2002).
Benner, S.A., Ellington, A.D. & Tauer, A. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. USA 86, 7054–7058 (1989).
Joyce, G.F. The antiquity of RNA-based evolution. Nature 418, 214–221 (2002).
White III, H.B. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 7, 101–104 (1976).
Jeffares, D.C., Poole, A.M. & Penny, D. Relics from the RNA world. J. Mol. Evol. 46, 18–36 (1998).
Grundy, F.J. & Henkin, T.M. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol. Microbiol. 30, 737–749 (1998).
Henkin, T.M. Transcription termination control in bacteria. Curr. Opin. Microbiol. 3, 149–153 (2000).
Grundy, F.J. & Henkin, T.M. The T box and S box transcription termination control systems. Frontiers Biosci. 8, D20–31 (2003).
Soukup, G.A. & Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999).
Gold, L., Poliski, B., Uhlenbeck, O.C. & Yarus, M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 64, 763–797 (1995).
Takusagawa, F., Fujioka, M., Spies, A. & Schowen, R.L. In Comprehensive Biological Catalysis Vol. 1 (ed. Sinnott, M.) 1–30 (Academic, New York, 1998).
McDaniel, B.A.M., Grundy, F.J., Artsimovitch, I. & Henkin, T.M. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100, 3083–3088 (2003).
Posnick, L.M. & Samson, L.D. Influence of S-adenosylmethionine pool size on spontaneous mutation, dam methylation, and cell growth of Escherichia coli. J. Bacteriol. 181, 6756–6762 (1999).
Epshtein, V., Mironov, A.S. & Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. USA 100, 5052–5056 (2003).
Mansilla, M.C., Albanesi, D. & De Mendoza, D. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in L-cysteine biosynthesis in Bacillus subtilis. J. Bacteriol. 182, 5885–5892 (2000).
Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C. & Breaker, R.R. Metabolite-sensing riboswitches control fundamental biochemical pathways in bacteria. Cell 113, 577–586 (2003).
Seetharaman, S., Zivartz, M., Sudarsan, N. & Breaker, R.R. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 19, 336–341 (2001).
Guérout-Fleury, A.M., Frandsen, N. & Stragier, P. Plasmids for ectopic integration in Bacillus subtilis. Gene 180, 57–61 (1996).
Fersht, A. In Structure and Mechanism in Protein Science 113 (Freeman, New York, 1999).
Jarmer, H., Berka, R., Knudsen, S. & Saxild, H.H. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol. Lett. 206, 197–200 (2002).
Acknowledgements
We thank J.K. Wickiser for assistance with the Scatchard analysis, other members of the Breaker laboratory for helpful discussions, and D. Söll at Yale University for the generous gift of B. subtilis strain 168. We also thank A. Miranker and B. Koo for assistance in confirming the purity of SAM using mass spectroscopy. This work was supported by grants from the US National Institutes of Health and the US National Science Foundation. R.R.B is also grateful for support from the David and Lucile Packard Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Winkler, W., Nahvi, A., Sudarsan, N. et al. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Mol Biol 10, 701–707 (2003). https://doi.org/10.1038/nsb967
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb967
This article is cited by
-
Magnesium ions mediate ligand binding and conformational transition of the SAM/SAH riboswitch
Communications Biology (2023)
-
Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution
Nature Communications (2023)
-
Structure-based insights into recognition and regulation of SAM-sensing riboswitches
Science China Life Sciences (2023)
-
Getting to the bottom of lncRNA mechanism: structure–function relationships
Mammalian Genome (2022)
-
Identification of 11 candidate structured noncoding RNA motifs in humans by comparative genomics
BMC Genomics (2021)