Abstract
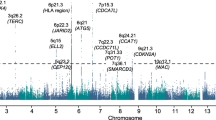
To identify risk variants for multiple myeloma, we conducted a genome-wide association study of 1,675 individuals with multiple myeloma and 5,903 control subjects. We identified risk loci for multiple myeloma at 3p22.1 (rs1052501 in ULK4; odds ratio (OR) = 1.32; P = 7.47 × 10−9) and 7p15.3 (rs4487645, OR = 1.38; P = 3.33 × 10−15). In addition, we observed a promising association at 2p23.3 (rs6746082, OR = 1.29; P = 1.22 × 10−7). Our study identifies new genomic regions associated with multiple myeloma risk that may lead to new etiological insights.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Kyle, R.A. & Rajkumar, S.V. Multiple myeloma. N. Engl. J. Med. 351, 1860–1873 (2004).
Palumbo, A. & Anderson, K. Multiple myeloma. N. Engl. J. Med. 364, 1046–1060 (2011).
Kyle, R.A. et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N. Engl. J. Med. 346, 564–569 (2002).
Landgren, O. et al. Risk of plasma cell and lymphoproliferative disorders among 14621 first-degree relatives of 4458 patients with monoclonal gammopathy of undetermined significance in Sweden. Blood 114, 791–795 (2009).
Altieri, A., Chen, B., Bermejo, J.L., Castro, F. & Hemminki, K. Familial risks and temporal incidence trends of multiple myeloma. Eur. J. Cancer 42, 1661–1670 (2006).
Morgan, G.J. et al. First-line treatment with zoledronic acid as compared with clodronic acid in multiple myeloma (MRC Myeloma IX): a randomised controlled trial. Lancet 376, 1989–1999 (2010).
Power, C. & Elliott, J. Cohort profile: 1958 British birth cohort (National Child Development Study). Int. J. Epidemiol. 35, 34–41 (2006).
Schmermund, A. et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL Study. Risk Factors, Evaluation of Coronary Calcium and Lifestyle. Am. Heart J. 144, 212–218 (2002).
Clayton, D.G. et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 37, 1243–1246 (2005).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Pettiti, D. Meta-analysis Decision Analysis and Cost-Effectiveness Analysis (Oxford University Press, New York, USA, 1994).
Tomlinson, I.P. et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 40, 623–630 (2008).
Jung, C.H., Ro, S.H., Cao, J., Otto, N.M. & Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 (2010).
Liang, C. & Jung, J.U. Autophagy genes as tumor suppressors. Curr. Opin. Cell Biol. 22, 226–233 (2010).
Ocio, E.M., Mateos, M.V., Maiso, P., Pandiella, A. & San-Miguel, J.F. New drugs in multiple myeloma: mechanisms of action and phase I/II clinical findings. Lancet Oncol. 9, 1157–1165 (2008).
Taddesse-Heath, L., Meloni-Ehrig, A., Scheerle, J., Kelly, J.C. & Jaffe, E.S. Plasmablastic lymphoma with MYC translocation: evidence for a common pathway in the generation of plasmablastic features. Mod. Pathol. 23, 991–999 (2010).
Dib, A., Gabrea, A., Glebov, O.K., Bergsagel, P.L. & Kuehl, W.M. Characterization of MYC translocations in multiple myeloma cell lines. J. Natl. Cancer Inst. Monogr. 39, 25–31 (2008).
Huang, A. et al. Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 65, 5607–5619 (2005).
Maertens, G.N., Cherepanov, P. & Engelman, A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 119, 2563–2571 (2006).
Blake, D.J. et al. β-dystrobrevin, a member of the dystophin-related protein family. Proc. Natl. Acad. Sci. USA 95, 241–246 (1998).
Matys, V. et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006).
Ferretti, V. et al. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 35, D122–D126 (2007).
Walker, B.A. et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 108, 1733–1743 (2006).
Stranger, B.E. et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1, e78 (2005).
Stranger, B.E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–853 (2007).
Fonseca, R. et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia 23, 2210–2221 (2009).
Dewald, G.W., Kyle, R.A., Hicks, G.A. & Greipp, P.R. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood 66, 380–390 (1985).
Debes-Marun, C.S. et al. Chromosome abnormalities clustering and its implications for pathogenesis and prognosis in myeloma. Leukemia 17, 427–436 (2003).
Walker, B.A. et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 116, e56–e65 (2010).
Landgren, O. & Weiss, B.M. Patterns of monoclonal gammopathy of undetermined significance and multiple myeloma in various ethnic/racial groups: support for genetic factors in pathogenesis. Leukemia 23, 1691–1697 (2009).
Enciso-Mora, V. et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat. Genet. 42, 1126–1130 (2010).
Crowther-Swanepoel, D. et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 42, 132–136 (2010).
Chiecchio, L. et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia 20, 1610–1617 (2006).
Neben, K. et al. Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica 95, 1150–1157 (2010).
Wuilleme, S. et al. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia 19, 275–278 (2005).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Higgins, J.P. & Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Ioannidis, J.P., Ntzani, E.E. & Trikalinos, T.A. 'Racial' differences in genetic effects for complex diseases. Nat. Genet. 36, 1312–1318 (2004).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Houlston, R.S. & Ford, D. Genetics of coeliac disease. QJM 89, 737–743 (1996).
Myers, S., Bottolo, L., Freeman, C., McVean, G. & Donnelly, P. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Cuzick, J. A Wilcoxon-type test for trend. Stat. Med. 4, 87–90 (1985).
Acknowledgements
We are grateful to all study participants and investigators at the individual centers for taking part. We are extremely grateful to all investigators who contributed to the generation of this data set. We also thank the staff of the Clinical Trials Research Unit (CTRU), University of Leeds, and the National Cancer Research Institute (NCRI) Haematology Clinical Studies Group. We are grateful to all investigators who contributed to the Colorectal Tumour Gene Identification (CORGI) consortium, from which controls in the replication analysis were drawn. The current study made use of genotyping data on the 1958 Birth Cohort. Genotyping data for control subjects was generated by the Wellcome Trust Sanger Institute. A full list of the investigators who contributed to the generation of the data is available from the WTCCC website (see URLs). The GWAS made use of genotyping data from the population-based HNR study. The HNR study was supported by the Heinz-Nixdorf Foundation. The genotyping of the HNR subjects on Illumina HumanOmni1-Quad BeadChips was financed by the German Centre for Neurodegenerative Disorders (DZNE), Bonn. Myeloma UK provided the main funding for the study. Additional funding was provided by Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), the Leukaemia Lymphoma Research Fund and the National Health Services (NHS) through the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital NHS Trust. In Germany, funding was provided to Dietmar-Hopp-Stiftung in Walldorf, the University Hospital Heidelberg and a grant from APO-STS (European Union Health-F4-2007-200767). Additionally, the study was funded by the German Ministry of Education and Science and the German Research Council (DFG; Projects SI 236/8-1, SI236/9-1 and ER 155/6-1).
Author information
Authors and Affiliations
Contributions
R.S.H. designed the study. R.S.H. and G.J.M. obtained financial support in the UK, and K.H. and H.G. obtained funding in Germany. D.C. performed the main statistical and bioinformatic analyses, and Y.P.M. and S.E.D. performed additional related analyses. P.B. coordinated laboratory studies. A.L. and B.O. performed genotyping of the UK samples. P.H., T.W.M. and M.M.N. performed and coordinated genotyping of the German controls; K.H. and A.F. coordinated genotyping of the German cases. D.C.J. managed and prepared the Myeloma-VII and Myeloma-IX case study DNA samples. H.G., K.N. and N.W. coordinated and managed the German DNA samples. G.J.M., F.E.D., W.A.G., G.H.J. and J.A.C. ascertained and collected case study samples from the UK Myeloma-VII and Myeloma-IX studies. S.M. obtained and managed the HNR samples. I.P.T. acquired colorectal cancer control samples. B.A.W. performed expression analyses on the UK samples. F.M.R. performed FISH analyses on the UK samples, and A.J. performed these analyses on the German samples. R.S.H. drafted the manuscript, and all authors contributed to the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3 and Supplementary Figures 1–4. (PDF 5579 kb)
Rights and permissions
About this article
Cite this article
Broderick, P., Chubb, D., Johnson, D. et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat Genet 44, 58–61 (2012). https://doi.org/10.1038/ng.993
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.993
This article is cited by
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
A genetic risk score of alleles related to MGUS interacts with socioeconomic position in a population-based cohort
Scientific Reports (2022)
-
Functional dissection of inherited non-coding variation influencing multiple myeloma risk
Nature Communications (2022)
-
A polygenic risk score for multiple myeloma risk prediction
European Journal of Human Genetics (2022)
-
Genetically determined telomere length and multiple myeloma risk and outcome
Blood Cancer Journal (2021)