Abstract
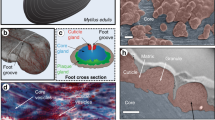
The beak of the jumbo squid Dosidicus gigas is a fascinating example of how seamlessly nature builds with mechanically mismatched materials. A 200-fold stiffness gradient begins in the hydrated chitin of the soft beak base and gradually increases to maximum stiffness in the dehydrated distal rostrum. Here, we combined RNA-Seq and proteomics to show that the beak contains two protein families. One family consists of chitin-binding proteins (DgCBPs) that physically join chitin chains, whereas the other family comprises highly modular histidine-rich proteins (DgHBPs). We propose that DgHBPs play multiple key roles during beak bioprocessing, first by forming concentrated coacervate solutions that diffuse into the DgCBP-chitin scaffold, and second by inducing crosslinking via an abundant GHG sequence motif. These processes generate spatially controlled desolvation, resulting in the impressive biomechanical gradient. Our findings provide novel molecular-scale strategies for designing functional gradient materials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Farren, L., Shayler, S. & Ennos, A.R. The fracture properties and mechanical design of human fingernails. J. Exp. Biol. 207, 735–741 (2004).
Benjamin, M. et al. Where tendons and ligaments meet bone: attachment sites ('entheses') in relation to exercise and/or mechanical load. J. Anat. 208, 471–490 (2006).
Lu, H.H. & Thomopoulos, S. Functional attachment of soft tissues to bone: development, healing, and tissue engineering. Annu. Rev. Biomed. Eng. 15, 201–226 (2013).
Zajac, F.E. Muscle and tendon: properties, models, scaling, and application to biomechanics and motor control. Crit. Rev. Biomed. Eng. 17, 359–411 (1989).
Sun, C.J. & Waite, J.H. Mapping chemical gradients within and along a fibrous structural tissue, mussel byssal threads. J. Biol. Chem. 280, 39332–39336 (2005).
Miserez, A., Schneberk, T., Sun, C., Zok, F.W. & Waite, J.H. The transition from stiff to compliant materials in squid beaks. Science 319, 1816–1819 (2008).
Suresh, S. Graded materials for resistance to contact deformation and damage. Science 292, 2447–2451 (2001).
Waite, J.H., Lichtenegger, H.C., Stucky, G.D. & Hansma, P. Exploring molecular and mechanical gradients in structural bioscaffolds. Biochemistry 43, 7653–7662 (2004).
Mikos, A.G. et al. Engineering complex tissues. Tissue Eng. 12, 3307–3339 (2006).
Helton, K.L., Ratner, B.D. & Wisniewski, N.A. Biomechanics of the sensor-tissue interface-effects of motion, pressure, and design on sensor performance and the foreign body response-part I: theoretical framework. J. Diabetes Sci. Technol. 5, 632–646 (2011).
Fleckman, P. & Olerud, J.E. Models for the histologic study of the skin interface with percutaneous biomaterials. Biomed. Mater. 3, 034006 (2008).
Fox, J.D., Capadona, J.R., Marasco, P.D. & Rowan, S.J. Bioinspired water-enhanced mechanical gradient nanocomposite films that mimic the architecture and properties of the squid beak. J. Am. Chem. Soc. 135, 5167–5174 (2013).
Zvarec, O., Purushotham, S., Masic, A., Ramanujan, R.V. & Miserez, A. Catechol-functionalized chitosan/iron oxide nanoparticle composite inspired by mussel thread coating and squid beak interfacial chemistry. Langmuir 29, 10899–10906 (2013).
Miserez, A., Rubin, D. & Waite, J.H. Cross-linking chemistry of squid beak. J. Biol. Chem. 285, 38115–38124 (2010).
Rubin, D.J., Miserez, A. & Waite, J.H. Diverse strategies of protein sclerotization in marine invertebrates: structure-property relationships in natural biomaterials. Adv. Insect Physiol. 38, 75–133 (2010).
Dilly, P.N. & Nixon, M. The cells that secrete the beaks in octopods and squids (Mollusca, Cephalopoda). Cell Tissue Res. 167, 229–241 (1976).
Grabherr, M.G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
de Kruif, C.G., Weinbreck, F. & De Vries, R.K. Complex coacervation of proteins and anionic polysaccharides. Curr. Opin. Colloid Interface Sci. 9, 340–349 (2004).
Smith, B.J. in The Protein Protocols Handbook (ed. Walker, J.M.) 507–510 (Humana Press Inc., 2002).
Miserez, A., Li, Y., Waite, J.H. & Zok, F. Jumbo squid beaks: inspiration for design of robust organic composites. Acta Biomater. 3, 139–149 (2007).
Sahal, D. et al. Specific and instantaneous one-step chemodetection of histidine-rich proteins by Pauly's stain. Anal. Biochem. 308, 405–408 (2002).
de Castro, E. et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 (2006).
Li, A.Q. et al. Chemical cleavage at aspartyl residues for protein identification. Anal. Chem. 73, 5395–5402 (2001).
Tan, Y., Yildiz, U.H., Wei, W., Waite, J.H. & Miserez, A. Layer-by-layer polyelectrolyte deposition: a mechanism for forming biocomposite materials. Biomacromolecules 14, 1715–1726 (2013).
Mithieux, S.M. & Weiss, A.S. in Fibrous Proteins: Coiled-Coils, Collagen and Elastomers (Advances in Protein Chemistry vol. 7) (eds. Parry, D.A.D. & Squire, J.M.) 437–461 (Elsevier Academic Press Inc., 2005).
Yeo, G.C., Keeley, F.W. & Weiss, A.S. Coacervation of tropoelastin. Adv. Colloid Interface Sci. 167, 94–103 (2011).
Wei, W. et al. A mussel-derived one component adhesive coacervate. Acta Biomater. 10, 1663–1670 (2014).
Ortony, J.H., Hwang, D.S., Franck, J.M., Waite, J.H. & Han, S. Asymmetric collapse in biomimetic complex coacervates revealed by local polymer and water dynamics. Biomacromolecules 14, 1395–1402 (2013).
Kizilay, E., Kayitmazer, A.B. & Dubin, P.L. Complexation and coacervation of polyelectrolytes with oppositely charged colloids. Adv. Colloid Interface Sci. 167, 24–37 (2011).
Betre, H., Setton, L.A., Meyer, D.E. & Chilkoti, A. Characterization of a genetically engineered elastin-like polypeptide for cartilaginous tissue repair. Biomacromolecules 3, 910–916 (2002).
Spruijt, E., Sprakel, J., Cohen-Stuart, M.A. & van der Gucht, J. Interfacial tension between a complex coacervate phase and its coexisting aqueous phase. Soft Matter 6, 172–178 (2010).
Weinbreck, F., de Vries, R., Schrooyen, P. & de Kruif, C.G. Complex coacervation of whey proteins and gum arabic. Biomacromolecules 4, 293–303 (2003).
Kaibara, K., Okazaki, T., Bohidar, H.B. & Dubin, P.L. pH-induced coacervation in complexes of bovine serum albumin and cationic polyelectrolytes. Biomacromolecules 1, 100–107 (2000).
Lin, J. & Brown, C.W. Spectroscopic measurement of NaCl and seawater salinity in the near-IR region of 680–1230 nm. Appl. Spectrosc. 47, 239–241 (1993).
Liu, B., Burdine, L. & Kodadek, T. Chemistry of periodate-mediated cross-linking of 3,4-dihydroxylphenylalanine-containing molecules to proteins. J. Am. Chem. Soc. 128, 15228–15235 (2006).
Kerwin, J.L. et al. Mass spectrometric analysis of catechol-histidine adducts from insect cuticle. Anal. Biochem. 268, 229–237 (1999).
Guerette, P.A. et al. Accelerating the design of biomimetic materials by integrating RNA-seq with proteomics and materials science. Nat. Biotechnol. 31, 908–915 (2013).
Zhao, H., Sun, C., Stewart, R.J. & Waite, J.H. Cement proteins of the tube-building polychaete Phragmatopoma californica. J. Biol. Chem. 280, 42938–42944 (2005).
Oparin, A.I. & Synge, A. The Origin of Life on the Earth 3rd edn. (Academic Press, 1957).
Hwang, D.S. et al. Viscosity and interfacial properties in a mussel-inspired adhesive coacervate. Soft Matter 6, 3232–3236 (2010).
Fournier, R.L. Basic Transport Phenomena in Biomedical Engineering 3rd edn. (CRC Press, Taylor & Francis Group, 2012).
Vrhovski, B. & Weiss, A.S. Biochemistry of tropoelastin. Eur. J. Biochem. 258, 1–18 (1998).
Giesa, T. & Buehler, M.J. Nanoconfinement and the strength of biopolymers. Annu. Rev. Biophys. 42, 651–673 (2013).
Gosline, J.M., Guerette, P.A., Ortlepp, C.S. & Savage, K.N. The mechanical design of spider silks: from fibroin sequence to mechanical function. J. Exp. Biol. 202, 3295–3303 (1999).
d'Orbigny, A.D. et al. Voyage dans l'Amérique méridionale: Tome Neuviéme (Pitois-Levrault, Paris, 1847).
Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 (2006).
Paz, M.A., Fluckiger, R., Boak, A., Kagan, H.M. & Gallop, P.M. Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 266, 689–692 (1991).
Stajich, J.E. et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12, 1611–1618 (2002).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J.V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Waterhouse, A.M., Procter, J.B., Martin, D.M.A., Clamp, M. & Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Newman, A.M. & Cooper, J.B. XSTREAM: A practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics 8, 382 (2007).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Rebers, J.E. & Willis, J.H. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem. Mol. Biol. 31, 1083–1093 (2001).
Goodarzi, M.T., Fotinopoulou, A. & Turner, G.A.A. in The Protein Protocols Handbook Springer Protocols Handbooks (ed. Walker, J.M.) 1205–1214 (Humana Press, 2009).
Hwang, D.S., Waite, J.H. & Tirrell, M. Promotion of osteoblast proliferation on complex coacervation-based hyaluronic acid—recombinant mussel adhesive protein coatings on titanium. Biomaterials 31, 1080–1084 (2010).
Acknowledgements
The authors would like to thank J. Pavlovich of the UCSB Mass Spectrometry Facility for his help with obtaining nanoLC-MS/MS data. We thank D. DeMartini (UCSB) and the crew of the R/V New Horizon for obtaining squid beak samples from the Guayamas Basin (Mexico), and K.W. Kong and V. Gangu (Molecular Engineering Lab, Agency for Science, Technology and Research (A*STAR), Singapore) for technical assistance with the RNA-Seq analysis. Some of the work was conducted in the Biological NanoStructures Laboratory within the California NanoSystems Institute, supported by the University California, Santa Barbara, and the University of California, Office of the President. This work was supported in whole or in part by US National Institutes of Health Grant R01-DE018468 and the Materials Research Science & Engineering Centers (MRSEC) Program of the US National Science Foundation under Award No. DMR 1121053 (J.H.W.). A.M. was supported by the Singapore National Research Foundation (NRF) through a NRF Fellowship, and S.H. by the Biomedical Research Council, A*STAR, Singapore.
Author information
Authors and Affiliations
Contributions
Y.T. carried out beak protein extraction and MS/MS experiments and prepared figures and tables. P.A.G. extracted and purified RNA samples, conducted 3′ RACE and designed DgHBP-1 cloning, expression and purification experiments. S.H. performed and analyzed RNA-Seq, did transcriptome analysis and RACE sequence confirmation and prepared figures. P.A.G., S.H., Y.T. and A.M. analyzed protein sequence data. C.H. expressed, purified and analyzed DgHBP-1. A.G. helped to purify DgHBP-1 and conducted rheological experiments. W.W. designed and carried out coacervation and infiltration experiments on recDgHBP-1. A.G. and C.H. conducted the characterization and infiltration of coacervates under the supervision of P.A.G. and A.M. Y.T. and W.W. carried out and analyzed coacervation and crosslinking experiments on DgHBP-pep. Y.T., J.H.W., P.A.G., S.H. and A.M. wrote the manuscript. J.H.W. and A.M. designed and supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–6 and Supplementary Figures 1–10. (PDF 15591 kb)
Coacervation of recDgHBP-1.
Part 1: Formation of a coacervate phase when a dilute solution of recDgHBP-1 is added to a 100 mM PBS buffer (pH 6.5), with a protein:buffer volume ratio of 1:9. Part 2: Dispersion of the coacervate phase by stirring. (MP4 14095 kb)
Rights and permissions
About this article
Cite this article
Tan, Y., Hoon, S., Guerette, P. et al. Infiltration of chitin by protein coacervates defines the squid beak mechanical gradient. Nat Chem Biol 11, 488–495 (2015). https://doi.org/10.1038/nchembio.1833
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1833
This article is cited by
-
Tuning the viscoelastic properties of peptide coacervates by single amino acid mutations and salt kosmotropicity
Communications Chemistry (2024)
-
Hierarchically structured bioinspired nanocomposites
Nature Materials (2023)
-
Liquid sculpture and curing of bio-inspired polyelectrolyte aqueous two-phase systems
Nature Communications (2023)
-
Fluid protein condensates for bio-inspired applications
Nature Reviews Bioengineering (2023)
-
Age and growth estimation of Southern Ocean squid Moroteuthopsis longimana: can we use beaks collected from predators’ stomachs?
Marine Biology (2023)