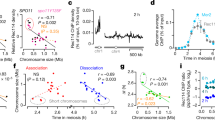
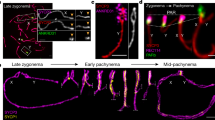
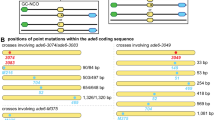
Abstract
During meiosis, accurate separation of maternal and paternal chromosomes requires that they first be connected to one another through homologous recombination. Meiotic recombination has many intriguing but poorly understood features that distinguish it from recombination in mitotically dividing cells, and several of these features depend on the meiosis-specific DNA strand exchange protein Dmc1 (disrupted meiotic cDNA1). Many questions about this protein have arisen since its discovery more than a decade ago, but recent genetic and biochemical breakthroughs promise to shed light on the unique behaviours and functions of this central player in the remarkable chromosome dynamics of meiosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Petronczki, M., Siomos, M. F. & Nasmyth, K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112, 423–440 (2003)
Kleckner, N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194 (2006)
Keeney, S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53 (2001)
Bianco, P. R., Tracy, R. B. & Kowalczykowski, S. C. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 3, D570–D603 (1998)
Shinohara, A. & Shinohara, M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet. Genome Res. 107, 201–207 (2004)
Zickler, D. & Kleckner, N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999)
Johnson, R. D. & Jasin, M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 29, 196–201 (2001)
Tarsounas, M., Morita, T., Pearlman, R. E. & Moens, P. B. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147, 207–220 (1999)
Bishop, D. K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092 (1994)
Bishop, D. K., Park, D., Xu, L. & Kleckner, N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456 (1992)
Shinohara, A., Ogawa, H. & Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–470 (1992)
Aboussekhra, A., Chanet, R., Adjiri, A. & Fabre, F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12, 3224–3234 (1992)
Krogh, B. O. & Symington, L. S. Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 (2004)
Schwacha, A. & Kleckner, N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90, 1123–1135 (1997)
Hunter, N. & Kleckner, N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001)
Grushcow, J. M. et al. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153, 607–620 (1999)
Stahl, F. W. et al. Does crossover interference count in Saccharomyces cerevisiae? Genetics 168, 35–48 (2004)
Shinohara, M., Sakai, K., Shinohara, A. & Bishop, D. K. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics 163, 1273–1286 (2003)
Tsubouchi, H. & Roeder, G. S. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell 5, 915–925 (2003)
Di Giacomo, M. et al. Distinct DNA damage-dependent and independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl Acad. Sci. USA 102, 737–742 (2005)
Rockmill, B., Sym, M., Scherthan, H. & Roeder, G. S. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 9, 2684–2695 (1995)
Young, J. A., Hyppa, R. W. & Smith, G. R. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167, 593–605 (2004)
Bishop, D. K. et al. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4, 425–444 (1999)
Shinohara, A., Gasior, S., Ogawa, T., Kleckner, N. & Bishop, D. K. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2, 615–629 (1997)
Paques, F. & Haber, J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 (1999)
Sehorn, M. G., Sigurdsson, S., Bussen, W., Unger, V. M. & Sung, P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429, 433–437 (2004)
Hong, E. L., Shinohara, A. & Bishop, D. K. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 276, 41906–41912 (2001)
Li, Z., Golub, E. I., Gupta, R. & Radding, C. M. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl Acad. Sci. USA 94, 11221–11226 (1997)
Masson, J. Y. et al. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 18, 6552–6560 (1999)
Nara, T., Hamada, F., Namekawa, S. & Sakaguchi, K. Strand exchange reaction in vitro and DNA-dependent ATPase activity of recombinant LIM15/DMC1 and RAD51 proteins from Coprinus cinereus. Biochem. Biophys. Res. Commun. 285, 92–97 (2001)
Passy, S. I. et al. Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl Acad. Sci. USA 96, 10684–10688 (1999)
Gupta, R. C., Golub, E., Bi, B. & Radding, C. M. The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc. Natl Acad. Sci. USA 98, 8433–8439 (2001)
Kinebuchi, T. et al. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol. Cell 14, 363–374 (2004)
Sauvageau, S. et al. Fission yeast Rad51 and Dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol. Cell. Biol. 25, 4377–4387 (2005)
Lee, M. H. et al. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J. Biol. Chem. 280, 40980–40984 (2005)
Bugreev, D. V., Golub, E. I., Stasiak, A. Z., Stasiak, A. & Mazin, A. V. Activation of human meiosis-specific recombinase Dmc1 by Ca2+. J. Biol. Chem. 280, 26886–26895 (2005)
Chang, Y. C. et al. Molecular visualization of the yeast Dmc1 protein ring and Dmc1–ssDNA nucleoprotein complex. Biochemistry 44, 6052–6058 (2005)
Kinebuchi, T., Kagawa, W., Kurumizaka, H. & Yokoyama, S. Role of the N-terminal domain of the human DMC1 protein in octamer formation and DNA binding. J. Biol. Chem. 280, 28382–28387 (2005)
Bugreev, D. V. & Mazin, A. V. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA 101, 9988–9993 (2004)
Chen, Y. K. et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl Acad. Sci. USA 101, 10572–10577 (2004)
Enomoto, R. et al. Positive role of the mammalian TBPIP/HOP2 protein in DMC1-mediated homologous pairing. J. Biol. Chem. 279, 35263–35272 (2004)
Petukhova, G. V. et al. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nature Struct. Mol. Biol. 12, 449–453 (2005)
Sung, P., Krejci, L., Van Komen, S. & Sehorn, M. G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278, 42729–42732 (2003)
Yang, H., Li, Q., Fan, J., Holloman, W. K. & Pavletich, N. P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature 433, 653–657 (2005)
Sharan, S. K. et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131, 131–142 (2004)
Siaud, N. et al. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23, 1392–1401 (2004)
Kojic, M., Kostrub, C. F., Buchman, A. R. & Holloman, W. K. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 10, 683–691 (2002)
Martin, J. S., Winkelmann, N., Petalcorin, M. I., McIlwraith, M. J. & Boulton, S. J. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 25, 3127–3139 (2005)
Dray, E., Siaud, N., Dubois, E. & Doutriaux, M. P. Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 140, 1059–1069 (2006)
Shinohara, M. et al. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147, 1545–1556 (1997)
Shinohara, M., Gasior, S. L., Bishop, D. K. & Shinohara, A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc. Natl Acad. Sci. USA 97, 10814–10819 (2000)
Petukhova, G., Stratton, S. & Sung, P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393, 91–94 (1998)
Petukhova, G., Sung, P. & Klein, H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14, 2206–2215 (2000)
Dresser, M. E. et al. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147, 533–544 (1997)
Catlett, M. G. & Forsburg, S. L. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol. Biol. Cell 14, 4707–4720 (2003)
Wesoly, J. et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 26, 976–989 (2006)
Van Komen, S., Petukhova, G., Sigurdsson, S., Stratton, S. & Sung, P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6, 563–572 (2000)
Ristic, D., Wyman, C., Paulusma, C. & Kanaar, R. The architecture of the human Rad54–DNA complex provides evidence for protein translocation along DNA. Proc. Natl Acad. Sci. USA 98, 8454–8460 (2001)
Jaskelioff, M., Van Komen, S., Krebs, J. E., Sung, P. & Peterson, C. L. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278, 9212–9218 (2003)
Alexeev, A., Mazin, A. & Kowalczykowski, S. C. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51–ssDNA nucleoprotein filament. Nature Struct. Biol. 10, 182–186 (2003)
Solinger, J. A., Kiianitsa, K. & Heyer, W. D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10, 1175–1188 (2002)
Leu, J. Y., Chua, P. R. & Roeder, G. S. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94, 375–386 (1998)
Gerton, J. L. & DeRisi, J. L. Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc. Natl Acad. Sci. USA 99, 6895–6900 (2002)
Petukhova, G. V., Romanienko, P. J. & Camerini-Otero, R. D. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5, 927–936 (2003)
Tsubouchi, H. & Roeder, G. S. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 22, 3078–3088 (2002)
Zierhut, C., Berlinger, M., Rupp, C., Shinohara, A. & Klein, F. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 14, 752–762 (2004)
Henry, J. M. et al. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol. Cell. Biol. 26, 2913–2923 (2006)
Enomoto, R. et al. Stimulation of DNA strand exchange by the human TBPIP/Hop2–Mnd1 complex. J. Biol. Chem. 281, 5575–5581 (2006)
Hayase, A. et al. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell 119, 927–940 (2004)
Tsubouchi, H. & Roeder, G. S. The budding yeast Mei5 and Sae3 proteins act together with Dmc1 during meiotic recombination. Genetics 168, 1219–1230 (2004)
Akamatsu, Y., Dziadkowiec, D., Ikeguchi, M., Shinagawa, H. & Iwasaki, H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl Acad. Sci. USA 100, 15770–15775 (2003)
Ellermeier, C., Schmidt, H. & Smith, G. R. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics 168, 1891–1898 (2004)
Neale, M. J., Pan, J. & Keeney, S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057 (2005)
Blat, Y., Protacio, R. U., Hunter, N. & Kleckner, N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802 (2002)
Smith, A. V. & Roeder, G. S. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136, 957–967 (1997)
Klein, F. et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 (1999)
Xu, H., Beasley, M. D., Warren, W. D., van der Horst, G. T. & McKay, M. J. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8, 949–961 (2005)
Acknowledgements
Work from the authors' laboratory is supported by a grant from the US National Institutes of Health (to S.K.). M.J.N. is supported in part by a fellowship from the Human Frontiers Science Program. S.K. is a Leukemia and Lymphoma Society Scholar.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Neale, M., Keeney, S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442, 153–158 (2006). https://doi.org/10.1038/nature04885
Issue Date:
DOI: https://doi.org/10.1038/nature04885
This article is cited by
-
Tracing the evolution of the plant meiotic molecular machinery
Plant Reproduction (2023)
-
A cytological revisit on parthenogenetic Artemia and the deficiency of a meiosis-specific recombinase DMC1 in the possible transition from bisexuality to parthenogenesis
Chromosoma (2023)
-
The Arabidopsis HOP2 gene has a role in preventing illegitimate connections between nonhomologous chromosome regions
Chromosome Research (2022)
-
Achiasmatic meiosis in the unisexual Amazon molly, Poecilia formosa
Chromosome Research (2022)
-
The genome loading model for the origin and maintenance of sex in eukaryotes
Biologia Futura (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.