Abstract
Although mutations in mitochondrial tRNAs constitute the most common mtDNA defect, the presence of pathological variants in mitochondrial tRNAAsn is extremely rare. We were able to identify a novel mtDNA tRNAAsn gene pathogenic mutation associated with a myopathic phenotype and a previously unreported respiratory impairment. Our proband is an adult woman with ophthalmoparesis and respiratory impairment. Her muscle biopsy presented several cytochrome c oxidase-negative (COX−) fibres and signs of mitochondrial proliferation (ragged red fibres). Sequence analysis of the muscle-derived mtDNA revealed an m.5709T>C substitution, affecting mitochondrial tRNAAsn gene. Restriction-fragment length polymorphism analysis of the mutation in isolated muscle fibres showed that a threshold of at least 91.9% mutated mtDNA results in the COX deficiency phenotype. The new phenotype further increases the clinical spectrum of mitochondrial diseases caused by mutations in the tRNAAsn gene.
Similar content being viewed by others
Introduction
Many mitochondrial disorders are associated with mutations in mitochondrial tRNAs. To date, over 200 point mutations affecting mitochondrial tRNA genes have been described.1
Nevertheless, mutations in the tRNAAsn gene are very rare; until now, only five pathogenic variants have been associated with clinical phenotypes ranging from chronic external ophthalmoplegia (cPEO), with or without mitochondrial myopathy, to lethal early-onset encephalomyopathy.2
Here we present clinical and molecular features of a patient with ophthalmoparesis and respiratory impairment associated with a novel heteroplasmic mutation (m.5709T>C) disclosed in the mitochondrial tRNAAsn gene.
Case report
The proband is a 51-year-old woman with a several year history of progressive external ophthalmoparesis (PEO) and bilateral eyelid ptosis, which was surgically corrected at age 29, but which gradually worsened again in the following years. At age 47 years, she was admitted to hospital for subacute onset of a severe respiratory insufficiency and she was diagnosed with a restrictive syndrome with indication for non-invasive ventilation (B-PAP) during the night. Her clinical history is complicated by hypothyroidism (she is taking replacement therapy) and anxious–depressive syndrome.
Neurological examination showed bilateral eyelid ptosis and bilateral, both vertical and horizontal, ophthalmoparesis without diplopia. She had mild axial and proximal upper limb weakness (bilateral sternocleidomastoid and deltoid muscles) with brisk tendon reflexes and no sensitive alterations. Neither cerebellar nor gait dysfunction were observed.
Blood tests were normal, except for mildly elevated CK (between 290 and 450 U/L, nv <185 U/L), LDH (872 U/L with nv 125–243 U/L) and basal lactate (3.9 mmol/l venous, with nv 0.90–1.70 mmol/l and 3.1 mmol/l arterial with nv 0.36–1.25 mmol/l).
EMG examination disclosed mild non-specific abnormalities in both deltoid and quadriceps muscles. Cardiological evaluation and tests were normal.
Respiratory functional tests confirmed a chronic respiratory insufficiency with restrictive syndrome.
Her younger sister, aged 42 years, has mild mental retardation and unspecified psychiatric disorders, but no eyelid ptosis or ophthalmoparesis. Her serum CK levels are normal. Both the mother and the maternal grandmother are reported affected with sarcoidosis and ophthalmoparesis (no clinical reports available). The father is healthy and neither the proband nor the sister has children.
At age 49 years, the patient underwent left deltoid muscle biopsy, which was consistent with a mitochondrial disorder.
Materials and methods
Histological and histochemical analysis of the muscle biopsy, Southern blot analysis of muscle mtDNA and PCR assay for multiple deletions were performed as described.3, 4, 5
Fragments encompassing the 22 tRNA genes were PCR-amplified, using a set of primers contained in the MitoSEQ Resequencing System (Applied Biosystem, Foster City, CA, USA) and sequenced on an Applied Biosystem 3100 Genetic Analyzer.
A 750-bp fragment encompassing m.5709T>C mutation was PCR-amplified. The mutated nucleotide C introduces a BstXI-restriction site. BstXI cuts the mutated molecules into two fragments of 500 and 250 bp, whereas the wild-type PCR fragment remains uncut.
Single fibres were microdissected from 30-μm thick muscle sections using the Leica Laser Microdissection Microscope ASLMD (Leica Microsystems, Wetzlar, Germany) and were processed as described.6
Mutational load assessment in single fibres, muscle biopsy and blood was performed by last-cycle hot PCR7 followed by restriction-fragment length polymorphism (RFLP) analysis. Aliquots (20 μl) of PCR products were digested and electrophoresed in 5% non-denaturing acrylamide gel. The proportion of mutant mtDNA was evaluated by densitometry using the NIH ImageJ software (http://rsbweb.nih.gov/ij/).
Results
Morphological and histochemical investigations
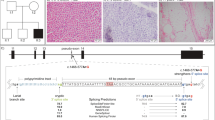
Left deltoid muscle biopsy showed well-preserved muscle architecture with evidence of ragged red fibres (RRFs; Figures 1a and c). Histochemical analysis revealed several cytochrome c oxidase (COX)-negative fibres (Figure 1b), many of them with increased SDH activity (Figure 1d) indicating mitochondrial proliferation.
Histological and histochemical findings in proband's muscle (a–d). (a) Original magnification × 200) and (c) × 400; Gomori trichrome histology showing RRFs (*) within a conserved muscle structure; (b, d × 100); COX (b) and combined COX-SDH (d) histochemistry showing severe and diffuse COX-deficiency with evidence of mitochondrial proliferation (*). (e) Cloverleaf structure of the human mitochondrial tRNAAsn (top); the m.5709T>C mutation resulting in A-to-G substitution at the RNA level is indicated by the arrow. Homologous sequences of part of the mitochondrial tRNAAsn in different species show a wide conservation of the affected nucleotide (bottom). All bases shown in black are 100% conserved in mammalian tRNAAsn genes. (f) A speculation of the mutated molecule secondary structure shows how it would likely result in a D-stem and -loop defective tRNA. (g) PCR–RFLP analysis of the m.5709T>C mutation. Autoradiogram showing BstXI-digested PCR products electrophoresed through a 5% non-denaturing polyacrylamide gel, performed on isolated fibres and on tissues from proband and controls. (h) Distribution of the mutation load measured in COX-positive (COX+, n=4) and COX-negative (COX−, n=5) muscle fibres after PCR–RFLP and densitometric analysis. Horizontal bars represent the calculated means.
Molecular genetic analysis
Multiple deletions of mtDNA were ruled out by Southern blot and PCR analyses.
Direct sequencing of PCR-amplified fragments encompassing the 22 tRNA genes of muscle mtDNA revealed a T-to-C transition at nucleotide position 5709. This substitution results in the replacement of a well-conserved A with a G in the cloverleaf secondary structure of the tRNAAsn gene, whose transcription proceeds on the opposite strand (Figure 1e). The A-to-G transition seems to result in a shift in the nucleotides involved in the D-stem formation, leading to an impaired D-stem and loop structure (Figure 1f). The mutation was not found in 100 Caucasian controls.
PCR–RFLP analysis showed that the mutation was heteroplasmic in skeletal muscle and in white blood cells, percentage of mutated genomes being 89.0% and 21.7%, respectively. The mutation was not detected in blood from the proband's sister (Figure 1g).
Single fibre analysis showed a higher degree of heteroplasmy in COX-deficient fibres (93.1±1.5%, n=5) compared with COX-positive muscle fibres (78.6±5.5%, n=4, Figures 1g and h). The difference between COX-positive and COX-negative fibres was statistically significant (P<0.01).
Discussion
More than half of the pathogenic mtDNA mutations are concentrated in tRNA genes.2 So far, five pathogenic point mutations within tRNAAsn gene have been described in seven probands: m.5703G>A associated with bilateral ptosis, cPEO and mitochondrial myopathy,8, 9 m.5692T>C associated with cPEO,10 m.5698G>A causing an isolated myopathy with cPEO in two sporadic patients,11, 12 m.5693T>C associated with a lethal early-onset encephalomyopathy13 and m.5728T>C leading to severe multiorgan failure.14
Here we report the novel substitution m.5709T>C in muscle-derived mtDNA from a woman affected with ophthalmoparesis and respiratory impairment. Our study meets most of the consensus criteria used to assign pathogenicity to mitochondrial tRNA variants:15 (i) m.5709T>C was found in heteroplasmic state; (ii) the proportion of mutated mtDNA was higher in the affected tissue (skeletal muscle) than in an unaffected tissue with rapid turnover (blood); (iii) the mutation occurs at an evolutionary conserved site in the mitochondrial genome and it seems to cause a significant structural impairment in the tRNAAsn D-stem and loop; (iv) m.5709T>C was absent in a cohort of ethnically matched healthy controls; (v) mutation load was significantly higher in isolated COX-negative fibres than in COX-positive single muscle fibres. We could not prove the matrilineal transmission, suggested by both the kind of mutation and the family history, as maternal mtDNA was unavailable. Muscle-derived mtDNA from the sister was not available to ascertain the presence of the mutation and, however, she does not have a myopathic phenotype. Because we did not detect mutated molecules in her leukocytes, we are currently unable to say whether the sister's clinical condition, mainly affecting the central nervous system, is another possible manifestation of the mutation-related clinical picture.
The small number of tRNAAsn mutations reported so far makes a genotype–phenotype correlation a hazardous task. Heterogeneous clinical presentations can be divided into two groups: an early onset syndromic disease leading to lethal mitochondrial encephalomyopathy13 or multi-organ failure,14 and an adult onset phenotype characterized by cPEO, myopathy and a neurological syndrome resembling MERRF (myoclonus epilepsy and RRF).8, 9, 10, 11, 12 As expected, mutations resulting in severe phenotypes or fatal outcome were present at almost homoplasmic levels in blood and skeletal muscle (>97%), whereas mutation load appeared lower in adult-onset patients, ranging from 46 to 80%. In our proband, the level of heteroplasmy in leukocytes was moderately low, whereas the mutation appeared largely heteroplasmic in skeletal muscle tissue. A narrow, although significant, mutational threshold could be still identified analysing isolated muscle fibres. Our patient shared the myopathic phenotype and developed cPEO, followed by a previously unreported respiratory impairment. Considering her sister's clinical features, an involvement of the central nervous system by the same mutation cannot be excluded.
Ventilatory failure is frequently described in both acute and chronic-progressive neuromuscular disorders.16 In the latter, it usually appears late in the course of the disease, but occasional presentation with respiratory failure has been reported, including restrictive syndrome in mitochondrial disorders.17 Two pathophysiological mechanisms leading to respiratory failure in patients with mitochondrial disorders have been proposed: (i) abnormal respiratory drive caused by dysfunction in the central nervous system (brainstem) respiratory centers; (ii) weakness and/or fatigue of the inspiratory muscles.
In our patient, the first mechanism is unlikely to be involved, because other clinical signs of brainstem dysfunction were absent. We therefore hypothesise an inspiratory muscle dysfunction due to both the fatigue from inappropriate energetic supply and from primary respiratory muscle weakness. Pulmonary function tests in our patient (single parameters not available) confirmed respiratory muscle weakness with evidence of a restrictive syndrome.
In conclusion, our data provide strong elements supporting the pathogenic role of the m.5709T>C mutation, and help define its threshold level. The new phenotype further increases the clinical spectrum of mitochondrial diseases caused by mutations in the tRNAAsn gene.
References
Scaglia F, Wong LJ : Human mitochondrial transfer RNAs: role of pathogenic mutation in disease. Muscle Nerve 2008; 37: 150–171.
Zifa E, Giannouli S, Theotokis P, Stamatis C, Mamuris Z, Stathopoulos C : Mitochondrial tRNA mutations: clinical and functional perturbations. RNA Biol 2007; 4: 38–66.
Sciacco M, Prelle A, Comi GP et al: Retrospective study of a large population of patients affected with mitochondrial disorders: clinical, morphological and molecular genetic evaluation. J Neurol 2001; 248: 778–788.
Zeviani M, Gellera C, Pannacci M et al: Tissue distribution and transmission of mitochondrial DNA deletions in mitochondrial myopathies. Ann Neurol 1990; 28: 94–97.
Moraes CT, Atencio DP, Oca-Cossio J, Diaz F : Techniques and pitfalls in the detection of pathogenic mitochondrial DNA mutations. J Mol Diagn 2003; 5: 197–208.
Sciacco M, Bonilla E, Schon EA, Di Mauro S, Moraes CT : Distribution of wild-type and common deletion form of mtDNA on normal and respiration deficient musclefibers from patients with mitochondrial myopathy. Hum Mol Genet 1994; 3: 13–19.
Moraes CT, Ricci E, Bonilla E, DiMauro S, Schon EA : The mitochondrial tRNA(Leu(UUR)) mutation in mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am J Hum Genet 1992; 50: 934–949.
Moraes CT, Ciacci F, Bonilla E et al: Two novel pathogenic mitochondrial DNA mutations affecting organelle number and protein synthesis. Is the tRNA(Leu(UUR)) gene an etiologic hot spot? J Clin Invest 1993; 92: 2906–2915.
Vives-Bauza C, Del Toro M, Solano A, Montoya J, Andreu AL, Roig M : Genotype-phenotype correlation in the 5703G>A mutation in the tRNA(ASN) gene of mitochondrial DNA. J Inherit Metab Dis 2003; 26: 507–508.
Seibel P, Lauber J, Klopstock T, Marsac C, Kadenbach B, Reichmann H : Chronic progressive external ophthalmoplegia is associated with a novel mutation in the mitochondrial tRNA(Asn) gene. Biochem Biophys Res Commun 1994; 204: 482–489.
Sternberg D, Chatzoglou E, Laforêt P et al: Mitochondrial DNA transfer RNA gene sequence variations in patients with mitochondrial disorders. Brain 2001; 124: 984–994.
Spinazzola A, Carrara F, Mora M, Zeviani M : Mitochondrial myopathy and ophthalmoplegia in a sporadic patient with the 5698G-->A mitochondrial DNA mutation. Neuromuscul Disord 2004; 14: 815–817.
Coulbault L, Herlicoviez D, Chapon F et al: A novel mutation in the mitochondrial tRNA Asn gene associated with a lethal disease. Biochem Biophys Res Commun 2005; 329: 1152–1154.
Meulemans A, Seneca S, Lagae L et al: A novel mitochondrial transfer RNA(Asn) mutation causing multiorgan failure. Arch Neurol 2006; 63: 1194–1198.
McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM : Assigning pathogenicity to mitochondrial tRNA mutations: when ‘definitely maybe’ is not good enough. Trends Genet 2004; 20: 591–596.
Hutchinson D, Whyte K : Neuromuscular disease and respiratory failure. Pract Neurol 2008; 8: 229–237.
Cros D, Palliyath S, DiMauro S, Ramirez C, Shamsnia M, Wizer B : Respiratory failure revealing mitochondrial myopathy in adults. Chest 1992; 101: 824–828.
Acknowledgements
Gratitude has to be expressed to the patient for participating in this research. We wish to thank especially the ‘Associazione Amici del Centro Dino Ferrari’ for their support. The financial support of the following research grant is gratefully acknowledged: Telethon – UILDM Project GUP09004 ‘Construction of a database for a nation-wide Italian collaborative network of mitochondrial diseases’, Associazione Amici del Centro Dino Ferrari, University of Milan, the Telethon project GTB07001, the Eurobiobank project QLTR-2001-02769 and RF 02.187 Criobanca Automatizzata di Materiale Biologico.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Ronchi, D., Sciacco, M., Bordoni, A. et al. The novel mitochondrial tRNAAsn gene mutation m.5709T>C produces ophthalmoparesis and respiratory impairment. Eur J Hum Genet 20, 357–360 (2012). https://doi.org/10.1038/ejhg.2011.238
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.238